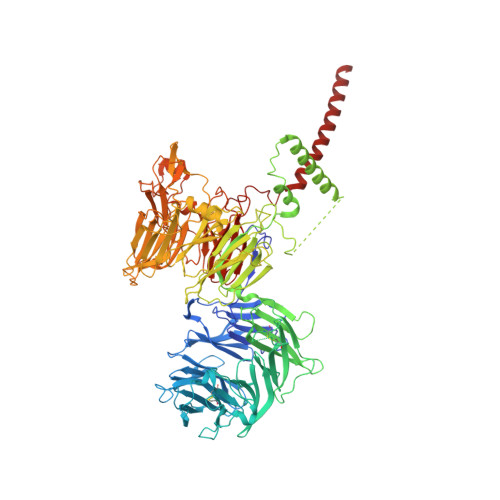
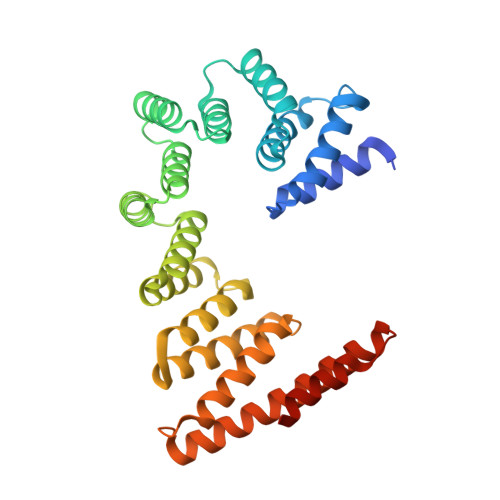
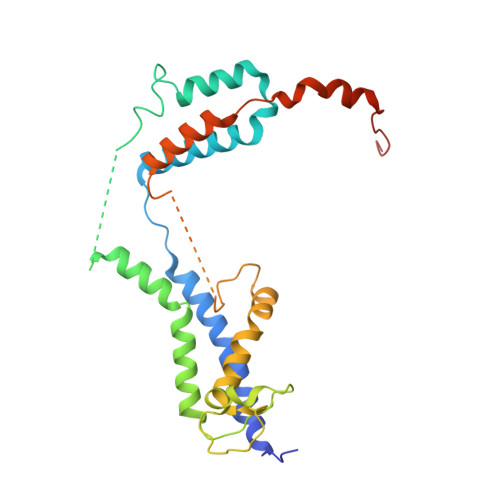
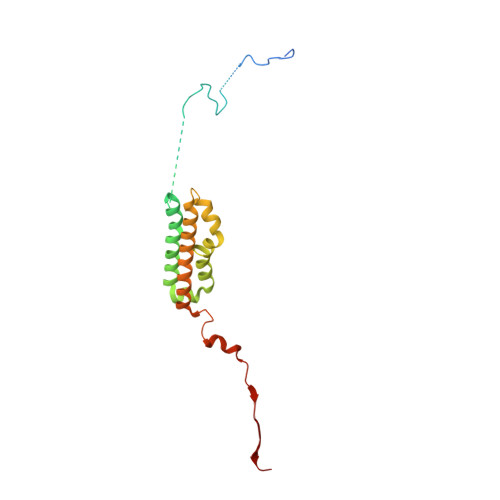
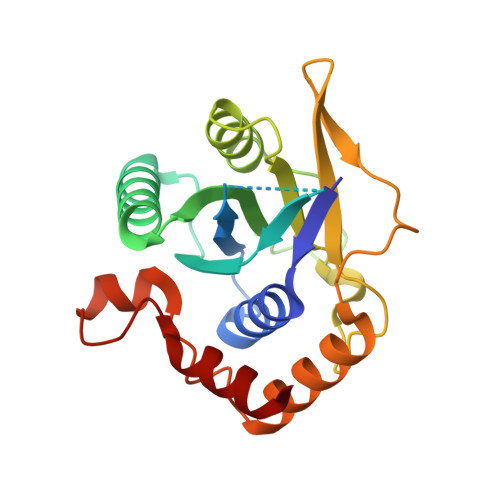
A selectivity filter in the ER membrane protein complex limits protein misinsertion at the ER.
Pleiner, T., Hazu, M., Pinton Tomaleri, G., Nguyen, V.N., Januszyk, K., Voorhees, R.M.(2023) J Cell Biol 222
- PubMed: 37199759
- DOI: https://doi.org/10.1083/jcb.202212007
- Primary Citation of Related Structures:
8S9S - PubMed Abstract:
Tail-anchored (TA) proteins play essential roles in mammalian cells, and their accurate localization is critical for proteostasis. Biophysical similarities lead to mistargeting of mitochondrial TA proteins to the ER, where they are delivered to the insertase, the ER membrane protein complex (EMC). Leveraging an improved structural model of the human EMC, we used mutagenesis and site-specific crosslinking to map the path of a TA protein from its cytosolic capture by methionine-rich loops to its membrane insertion through a hydrophilic vestibule. Positively charged residues at the entrance to the vestibule function as a selectivity filter that uses charge-repulsion to reject mitochondrial TA proteins. Similarly, this selectivity filter retains the positively charged soluble domains of multipass substrates in the cytosol, thereby ensuring they adopt the correct topology and enforcing the "positive-inside" rule. Substrate discrimination by the EMC provides a biochemical explanation for one role of charge in TA protein sorting and protects compartment integrity by limiting protein misinsertion.
- Division of Biology and Biological Engineering, California Institute of Technology , Pasadena, CA, USA.
Organizational Affiliation: