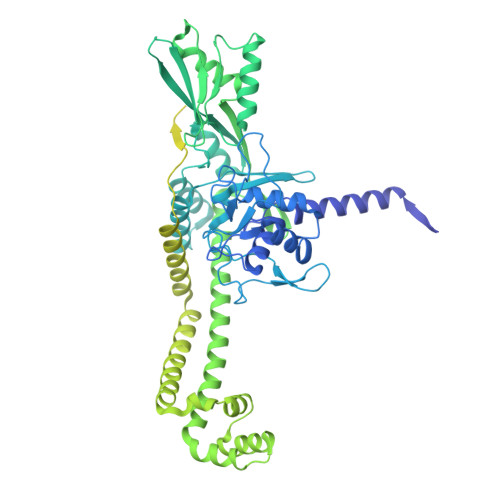
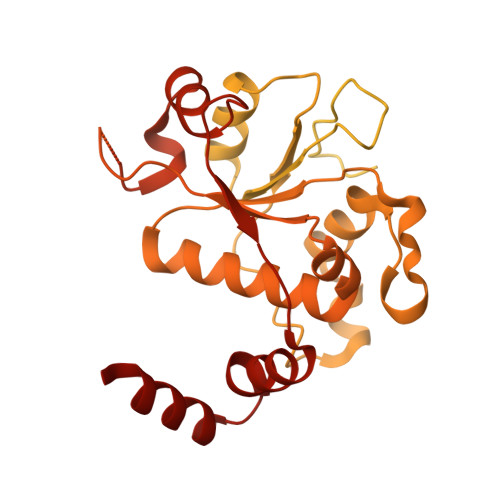
M. tuberculosis gyrase holocomplex with 150 bp DNA and gepotidacin
Gedeon, A., Yab, E., Dinut, A., Sadowski, E., Capton, E., Dreneau, A., Gioia, B., Piveteau, C., Djaout, K., Lecat, E., Wehenkel, A.M., Gubellini, F., Mechaly, A., Alzari, P.M., Deprez, B., Baulard, A., Aubry, A., Willand, N., Petrella, S.To be published.