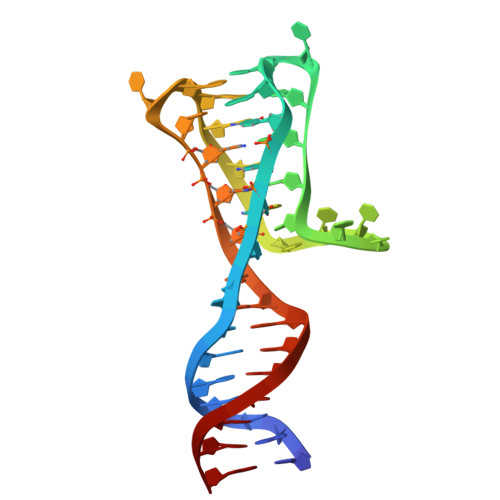
Unveiling the unusual i-motif-derived architecture of a DNA aptamer exhibiting high affinity for influenza A virus.
Tsvetkov, V., Mir, B., Alieva, R., Arutyunyan, A., Oleynikov, I., Novikov, R., Boravleva, E., Kamzeeva, P., Zatsepin, T., Aralov, A., Gonzalez, C., Zavyalova, E.(2025) Nucleic Acids Res 53
- PubMed: 39777463
- DOI: https://doi.org/10.1093/nar/gkae1282
- Primary Citation of Related Structures:
8S4N - PubMed Abstract:
Non-canonical nucleic acid structures play significant roles in cellular processes through selective interactions with proteins. While both natural and artificial G-quadruplexes have been extensively studied, the functions of i-motifs remain less understood. This study investigates the artificial aptamer BV42, which binds strongly to influenza A virus hemagglutinin and unexpectedly retains its i-motif structure even at neutral pH. However, BV42 conformational heterogeneity hinders detailed structural analysis. Molecular dynamics simulations and chemical modifications of BV42 helped us to identify a potential binding site, allowing for aptamer redesign to eliminate the conformational diversity while retaining binding affinity. Nuclear magnetic resonance spectroscopy confirmed the i-motif/duplex junction with the three-cytosine loop nearby. This study highlights the unique structural features of the functional i-motif and its role in molecular recognition of the target.
- Center for Mathematical Modeling in Drug Development, Sechenov First Moscow State Medical University, Moscow 119991, Russia.
Organizational Affiliation: