SilE-R and SilE-S-DABB Proteins Catalying Enantiospecific Hydrolysis of Organosilyl Ethers.
Pick, L.M., Oehme, V., Hartmann, J., Wenzlaff, J., Tang, Q., Grogan, G., Ansorge-Schumacher, M.B.(2024) Angew Chem Int Ed Engl 63: e202404105-e202404105
- PubMed: 38630059
- DOI: https://doi.org/10.1002/anie.202404105
- Primary Citation of Related Structures:
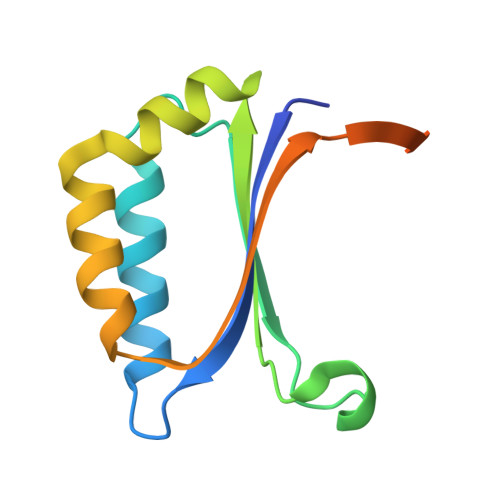
8S4E - PubMed Abstract:
Silyl ethers fulfil a fundamental role in synthetic organic chemistry as protecting groups and their selective cleavage is an important factor in their application. We present here for the first time two enzymes, SilE-R and SilE-S, which are able to hydrolyse silyl ethers. They belong to the stress-response dimeric A/B barrel domain (DABB) family and are able to cleave the Si-O bond with opposite enantiopreference. Silyl ethers containing aromatic, cyclic or aliphatic alcohols and, depending on the alcohol moiety, silyl functions as large as TBDMS are accepted. The X-ray crystal structure of SilE-R, determined to a resolution of 1.98 Å, in combination with mutational studies, revealed an active site featuring two histidine residues, H8 and H79, which likely act synergistically as nucleophile and Brønsted base in the hydrolytic mechanism, which has not previously been described for enzymes. Although the natural function of SilE-R and SilE-S is unknown, we propose that these 'silyl etherases' may have significant potential for synthetic applications.
- Professur für Molekulare Biotechnologie, Technische Universität Dresden, 01062, Dresden, Germany.
Organizational Affiliation: