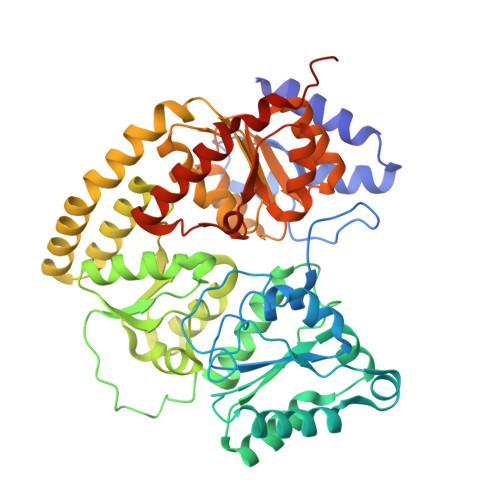
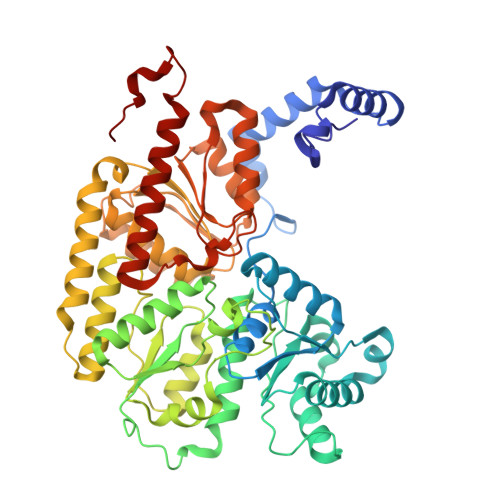
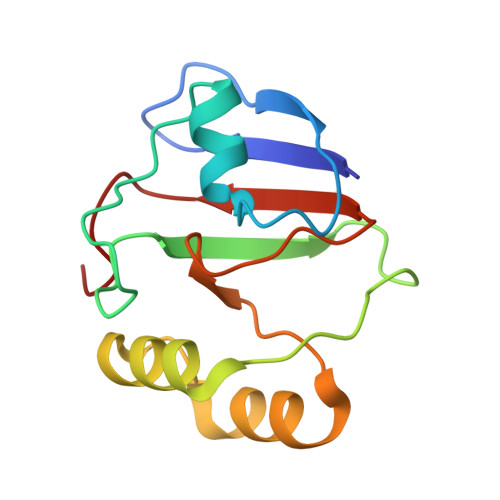
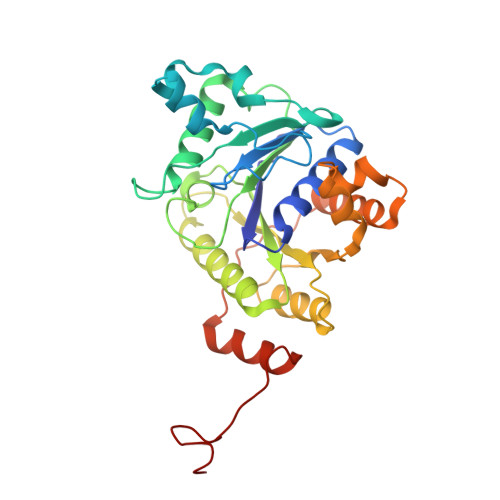
Conformational protection of molybdenum nitrogenase by Shethna protein II.
Franke, P., Freiberger, S., Zhang, L., Einsle, O.(2025) Nature 637: 998-1004
- PubMed: 39779845
- DOI: https://doi.org/10.1038/s41586-024-08355-3
- Primary Citation of Related Structures:
8RHO, 8RHP - PubMed Abstract:
The oxygen-sensitive molybdenum-dependent nitrogenase of Azotobacter vinelandii is protected from oxidative damage by a reversible 'switch-off' mechanism 1 . It forms a complex with a small ferredoxin, FeSII (ref. 2 ) or the 'Shethna protein II' 3 , which acts as an O 2 sensor and associates with the two component proteins of nitrogenase when its [2Fe:2S] cluster becomes oxidized 4,5 . Here we report the three-dimensional structure of the protective ternary complex of the catalytic subunit of Mo-nitrogenase, its cognate reductase and the FeSII protein, determined by single-particle cryo-electron microscopy. The dimeric FeSII protein associates with two copies of each component to assemble a 620 kDa core complex that then polymerizes into large, filamentous structures. This complex is catalytically inactive, but the enzyme components are quickly released and reactivated upon oxygen depletion. The first step in complex formation is the association of FeSII with the more O 2 -sensitive Fe protein component of nitrogenase during sudden oxidative stress. The action of this small ferredoxin represents a straightforward means of protection from O 2 that may be crucial for the maintenance of recombinant nitrogenase in food crops.
- Institut für Biochemie, Albert-Ludwigs-Universität Freiburg, Freiburg im Breisgau, Germany.
Organizational Affiliation: