Structure and tethering mechanism of dynein-2 intermediate chains in intraflagellar transport.
Mukhopadhyay, A.G., Toropova, K., Daly, L., Wells, J.N., Vuolo, L., Mladenov, M., Seda, M., Jenkins, D., Stephens, D.J., Roberts, A.J.(2024) EMBO J 43: 1257-1272
- PubMed: 38454149
- DOI: https://doi.org/10.1038/s44318-024-00060-1
- Primary Citation of Related Structures:
8RGG, 8RGH, 8RGI - PubMed Abstract:
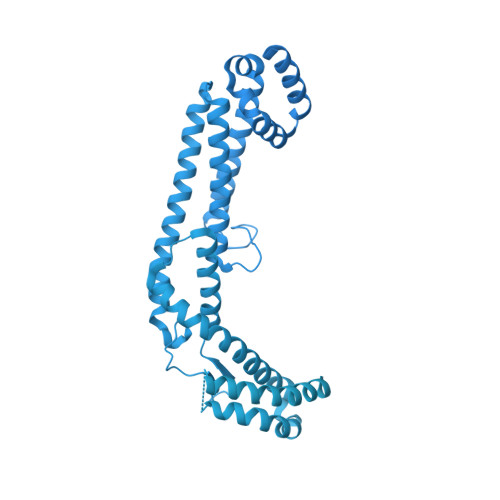
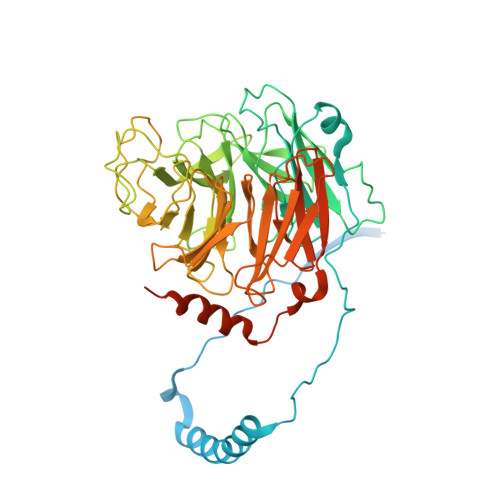
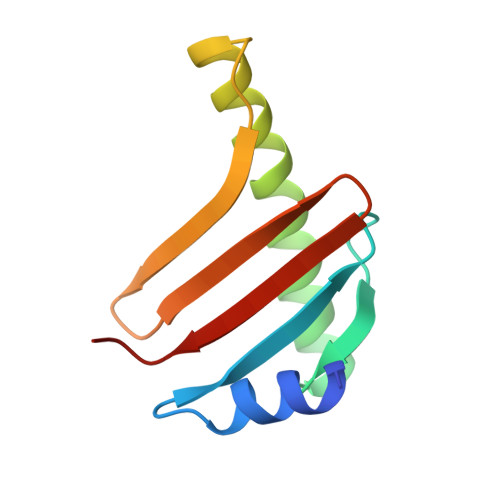
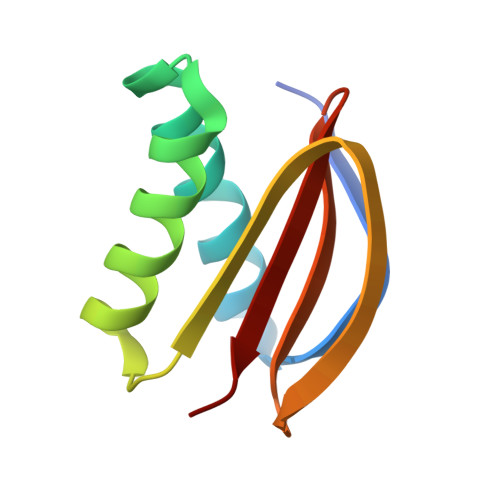
Dynein-2 is a large multiprotein complex that powers retrograde intraflagellar transport (IFT) of cargoes within cilia/flagella, but the molecular mechanism underlying this function is still emerging. Distinctively, dynein-2 contains two identical force-generating heavy chains that interact with two different intermediate chains (WDR34 and WDR60). Here, we dissect regulation of dynein-2 function by WDR34 and WDR60 using an integrative approach including cryo-electron microscopy and CRISPR/Cas9-enabled cell biology. A 3.9 Å resolution structure shows how WDR34 and WDR60 use surprisingly different interactions to engage equivalent sites of the two heavy chains. We show that cilia can assemble in the absence of either WDR34 or WDR60 individually, but not both subunits. Dynein-2-dependent distribution of cargoes depends more strongly on WDR60, because the unique N-terminal extension of WDR60 facilitates dynein-2 targeting to cilia. Strikingly, this N-terminal extension can be transplanted onto WDR34 and retain function, suggesting it acts as a flexible tether to the IFT "trains" that assemble at the ciliary base. We discuss how use of unstructured tethers represents an emerging theme in IFT train interactions.
- Sir William Dunn School of Pathology, University of Oxford, Oxford, UK.
Organizational Affiliation: