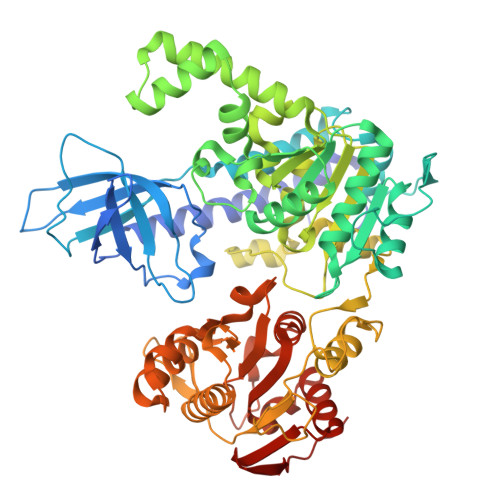
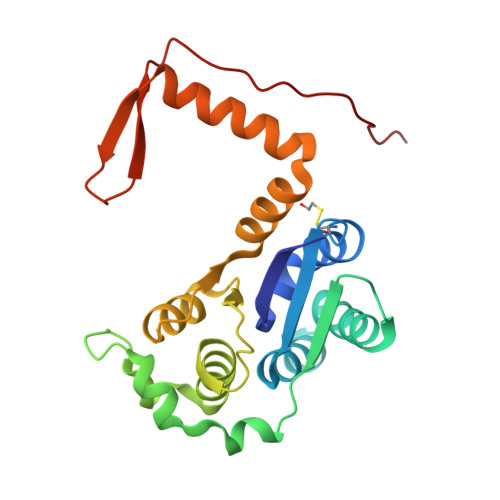
Structure of the Nmd4-Upf1 complex supports conservation of the nonsense-mediated mRNA decay pathway between yeast and humans.
Barbarin-Bocahu, I., Ulryck, N., Rigobert, A., Ruiz Gutierrez, N., Decourty, L., Raji, M., Garkhal, B., Le Hir, H., Saveanu, C., Graille, M.(2024) PLoS Biol 22: e3002821-e3002821
- PubMed: 39331656
- DOI: https://doi.org/10.1371/journal.pbio.3002821
- Primary Citation of Related Structures:
8RD3, 8RDD - PubMed Abstract:
The nonsense-mediated mRNA decay (NMD) pathway clears eukaryotic cells of mRNAs containing premature termination codons (PTCs) or normal stop codons located in specific contexts. It therefore plays an important role in gene expression regulation. The precise molecular mechanism of the NMD pathway has long been considered to differ substantially from yeast to metazoa, despite the involvement of universally conserved factors such as the central ATP-dependent RNA-helicase Upf1. Here, we describe the crystal structure of the yeast Upf1 bound to its recently identified but yet uncharacterized partner Nmd4, show that Nmd4 stimulates Upf1 ATPase activity and that this interaction contributes to the elimination of NMD substrates. We also demonstrate that a region of Nmd4 critical for the interaction with Upf1 in yeast is conserved in the metazoan SMG6 protein, another major NMD factor. We show that this conserved region is involved in the interaction of SMG6 with UPF1 and that mutations in this region affect the levels of endogenous human NMD substrates. Our results support the universal conservation of the NMD mechanism in eukaryotes.
- Laboratoire de Biologie Structurale de la Cellule (BIOC), CNRS, Ecole polytechnique, Institut Polytechnique de Paris, Palaiseau, France.
Organizational Affiliation: