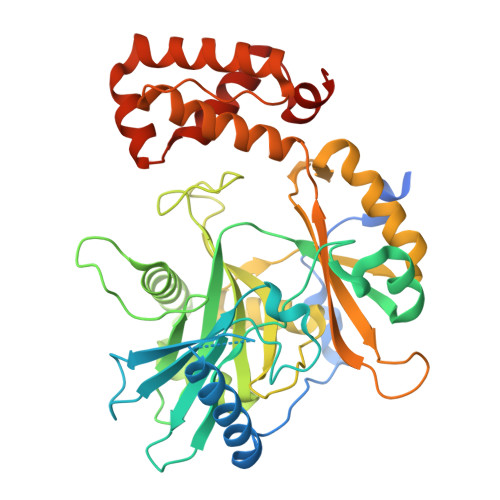
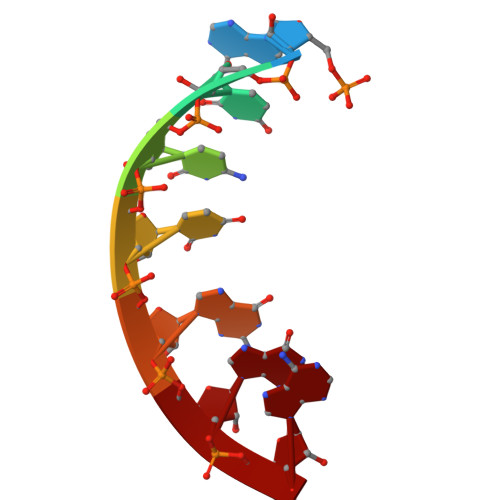
Structure of fungal tRNA ligase Trl1 with RNA reveals conserved substrate-binding principles.
Kohler, S., Kopp, J., Maiti, S., Bujnicki, J.M., Peschek, J.(2025) Nat Struct Mol Biol 32: 1657-1668
- PubMed: 40563009
- DOI: https://doi.org/10.1038/s41594-025-01589-3
- Primary Citation of Related Structures:
8RBJ - PubMed Abstract:
RNA ligases play a vital role in RNA processing and maturation, including tRNA splicing, RNA repair and the unfolded protein response (UPR). In fungi and plants, the tripartite tRNA ligase Trl1 catalyzes the joining of TSEN-cleaved pre-tRNA exon halves. Trl1 also functions as ligase in the non-conventional HAC1 mRNA splicing during the UPR. The final ligation step is performed by the N-terminal adenylyltransferase domain (ligase; LIG). The spatial arrangement of the exon ends during the ligation reaction has remained elusive. Here we report the crystal structure of Chaetomium thermophilum Trl1-LIG in complex with a tRNA-derived substrate. Our structure represents a snapshot of the activated RNA intermediate and defines the conserved substrate-binding interface. The underlying enzyme-substrate interplay reveals a substrate-binding principle shared by adenylyltransferases. Moreover, we identify the determinants of RNA end specificity as well as the specific roles of Trl1-LIG's subdomains during ligase activation, substrate binding and phosphoryl transfer.
- Heidelberg University, Biochemistry Center (BZH), Heidelberg, Germany.
Organizational Affiliation: