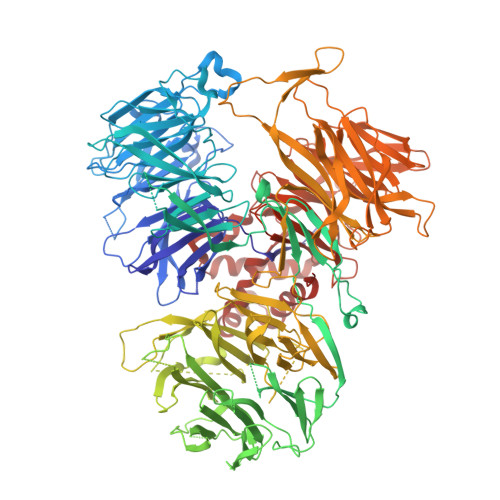
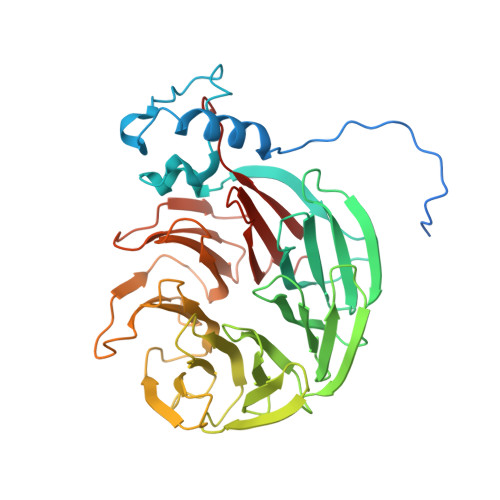
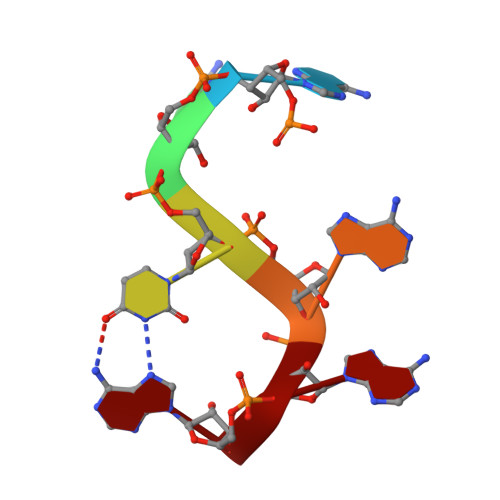
Molecular basis of human poly(A) polymerase recruitment by mPSF.
Todesca, S., Sandmeir, F., Keidel, A., Conti, E.(2024) RNA 30: 795-806
- PubMed: 38538052
- DOI: https://doi.org/10.1261/rna.079915.123
- Primary Citation of Related Structures:
8R8R - PubMed Abstract:
3' end processing of most eukaryotic pre-mRNAs is a crucial co-transcriptional process that generally involves the cleavage and polyadenylation of the precursor transcripts. Within the human 3' end processing machinery, the 4-subunit mammalian polyadenylation specificity factor (mPSF) recognizes the polyadenylation signal (PAS) in the pre-mRNA and recruits the polyA polymerase α (PAPOA) to it. To shed light on the molecular mechanisms of PAPOA recruitment to mPSF, we used a combination of cryogenic-electron microscopy (cryo-EM) single-particle analysis, computational structure prediction and in vitro biochemistry to reveal an intricate interaction network. A short linear motif in the mPSF subunit FIP1 interacts with the structured core of human PAPOA, with a binding mode that is evolutionary conserved from yeast to human. In higher eukaryotes, however, PAPOA contains a conserved C-terminal motif that can interact intramolecularly with the same residues of the PAPOA structured core used to bind FIP1. Interestingly, using biochemical assay and cryo-EM structural analysis, we found that the PAPOA C-terminal motif can also directly interact with mPSF at the subunit CPSF160. These results show that PAPOA recruitment to mPSF is mediated by two distinct intermolecular connections and further suggest the presence of mutually exclusive interactions in the regulation of 3' end processing.
- MPI of Biochemistry.
Organizational Affiliation: