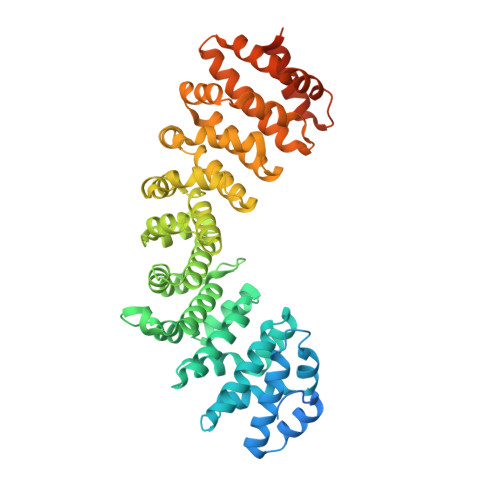
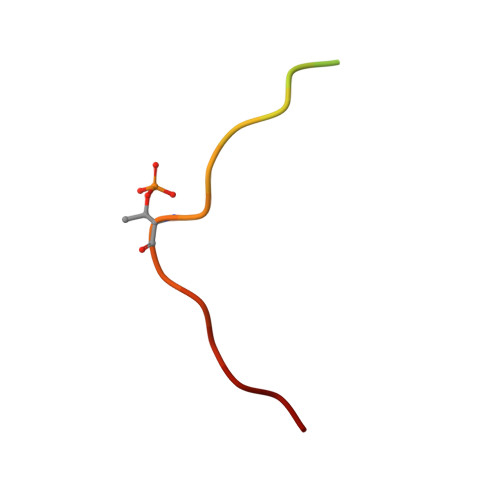
Structural determinants of phosphorylation-dependent nuclear transport of HCMV DNA polymerase processivity factor UL44.
Cross, E.M., Marin, O., Ariawan, D., Aragao, D., Cozza, G., Di Iorio, E., Forwood, J.K., Alvisi, G.(2024) FEBS Lett 598: 199-209
- PubMed: 38158756
- DOI: https://doi.org/10.1002/1873-3468.14797
- Primary Citation of Related Structures:
8QXW, 8QXX - PubMed Abstract:
Human cytomegalovirus DNA polymerase processivity factor UL44 is transported into the nucleus by importin (IMP) α/β through a classical nuclear localization signal (NLS), and this region is susceptible to cdc2-mediated phosphorylation at position T427. Whilst phosphorylation within and close to the UL44 NLS regulates nuclear transport, the details remain elusive, due to the paucity of structural information regarding the role of negatively charged cargo phosphate groups. We addressed this issue by studying the effect of UL44 T427 phosphorylation on interaction with several IMPα isoforms by biochemical and structural approaches. Phosphorylation decreased UL44/IMPα affinity 10-fold, and a comparative structural analysis of UL44 NLS phosphorylated and non-phosphorylated peptides complexed with mouse IMPα2 revealed the structural rearrangements responsible for phosphorylation-dependent inhibition of UL44 nuclear import.
- School of Dentistry and Medical Sciences, Charles Sturt University, Wagga Wagga, Australia.
Organizational Affiliation: