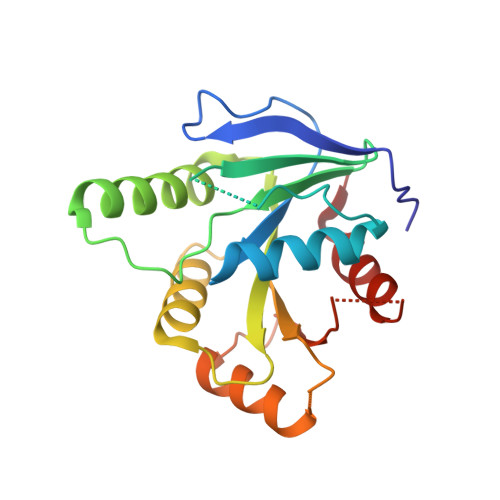
Crystal structure of GTPase YsxC from Staphylococcus aureus.
Biktimirov, A., Islamov, D., Fatkhullin, B., Lazarenko, V., Validov, S., Yusupov, M., Usachev, K.(2024) Biochem Biophys Res Commun 699: 149545-149545
- PubMed: 38277729
- DOI: https://doi.org/10.1016/j.bbrc.2024.149545
- Primary Citation of Related Structures:
8QUA - PubMed Abstract:
The YsxC protein from Staphylococcus aureus is a GTP-binding protein from the TRAFAC superfamily of the TrmE-Era-EngA-EngB-Septin-like GTPase class, EngB family of GTPases. Recent structural and biochemical studies of YsxC function show that it is an integral part of the pathogenic microorganism life cycle, as it is involved in the assembly of the large 50S ribosomal subunit. Structural studies of this protein with its specific functional features make it an attractive target for further development of new selective antimicrobials. In this study, we cloned the ysxC protein gene from S. aureus, overexpressed the protein in E. coli, and subsequently purified and crystallized it. Protein crystals were successfully grown using the vapor diffusion method, yielding diffraction data with a resolution of up to 2 Å. Comparative analysis of the structure of SaYsxC with known three-dimensional structures of homologs from other microorganisms showed the presence of structural differences for the apo form.
- Kazan Federal University, 18 Kremlyovskaya St., 420008, Kazan, Russian Federation.
Organizational Affiliation: