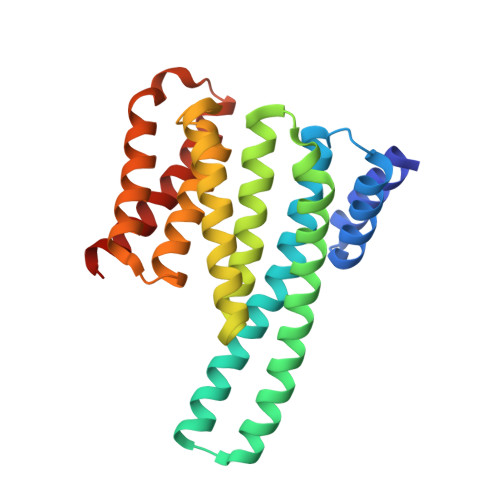
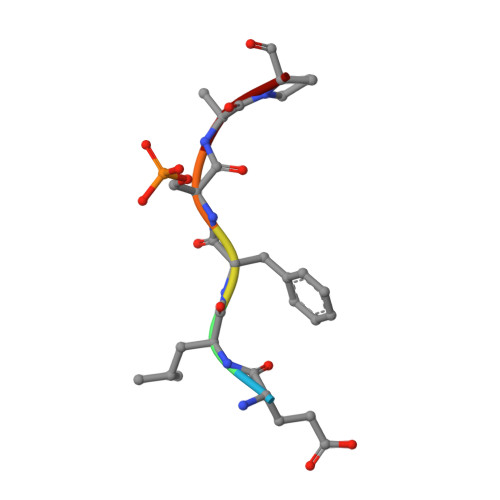
Mechanistic Insights into the Function of 14-3-3 Proteins as Negative Regulators of Brassinosteroid Signaling in Arabidopsis.
Obergfell, E., Hohmann, U., Moretti, A., Chen, H., Hothorn, M.(2024) Plant Cell Physiol 65: 1674-1688
- PubMed: 38783418
- DOI: https://doi.org/10.1093/pcp/pcae056
- Primary Citation of Related Structures:
8QT5, 8QTC, 8QTF, 8QTT - PubMed Abstract:
Brassinosteroids (BRs) are vital plant steroid hormones sensed at the cell surface by a membrane signaling complex comprising the receptor kinase BRI1 and a SERK-family co-receptor kinase. Activation of this complex lead to dissociation of the inhibitor protein BKI1 from the receptor and to differential phosphorylation of BZR1/BES1 transcription factors by the glycogen synthase kinase 3 protein BIN2. Many phosphoproteins of the BR signaling pathway, including BRI1, SERKs, BKI1 and BZR1/BES1 can associate with 14-3-3 proteins. In this study, we use quantitative ligand binding assays to define the minimal 14-3-3 binding sites in the N-terminal lobe of the BRI1 kinase domain, in BKI1, and in BZR1 from Arabidopsis thaliana. All three motifs require to be phosphorylated to specifically bind 14-3-3s with mid- to low micromolar affinity. BR signaling components display minimal isoform preference within the 14-3-3 non-ε subgroup. 14-3-3λ and 14-3-3ω isoform complex crystal structures reveal that BKI1 and BZR1 bind as canonical type II 14-3-3 linear motifs. Disruption of key amino acids in the phosphopeptide binding site through mutation impairs the interaction of 14-3-3λ with all three linear motifs. Notably, quadruple loss-of-function mutants from the non-ε group exhibit gain-of-function brassinosteroid signaling phenotypes, suggesting a role for 14-3-3 proteins as overall negative regulators of the BR pathway. Collectively, our work provides further mechanistic and genetic evidence for the regulatory role of 14-3-3 proteins at various stages of the brassinosteroid signaling cascade.
- Structural Plant Biology Laboratory, Department of Plant Sciences, University of Geneva, 1211 Geneva, Switzerland.
Organizational Affiliation: