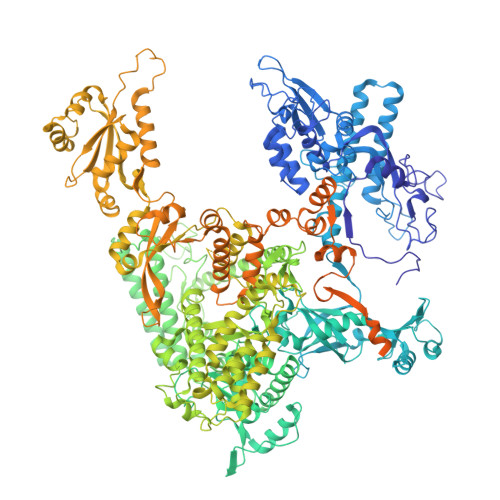
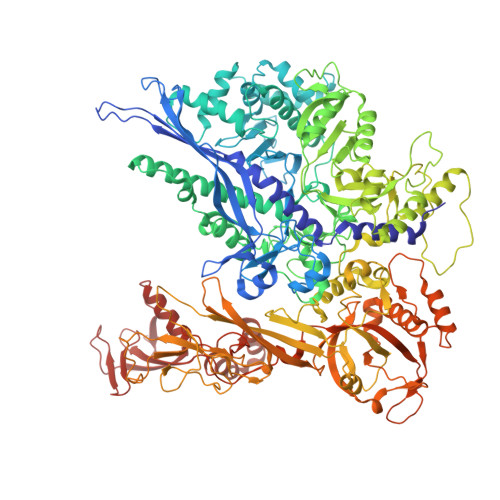
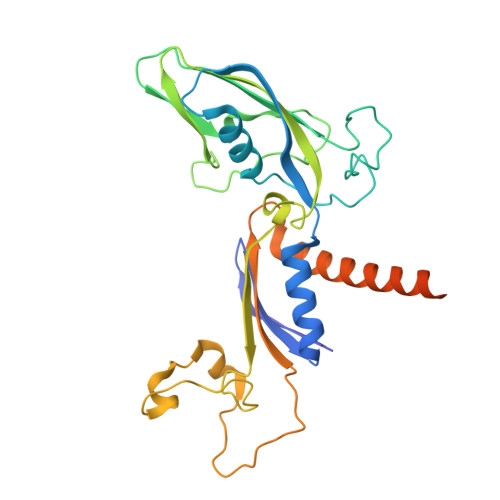
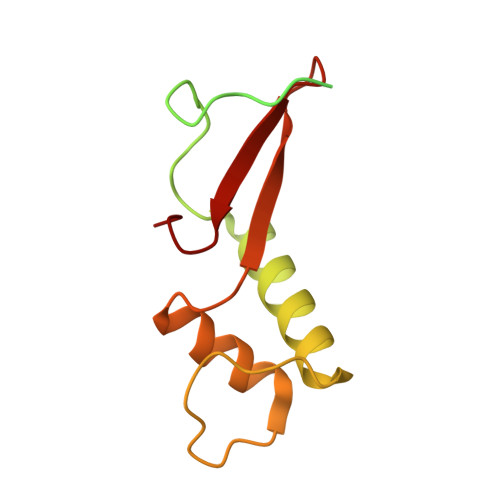
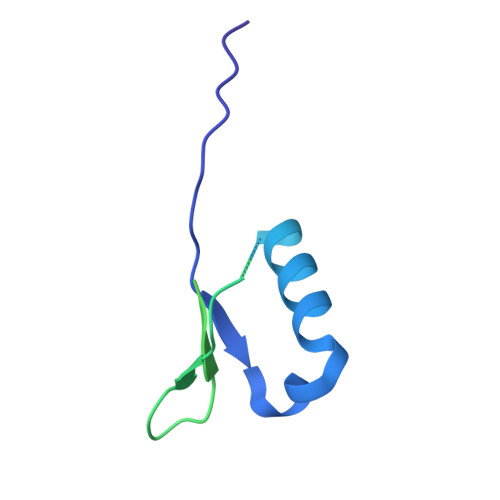
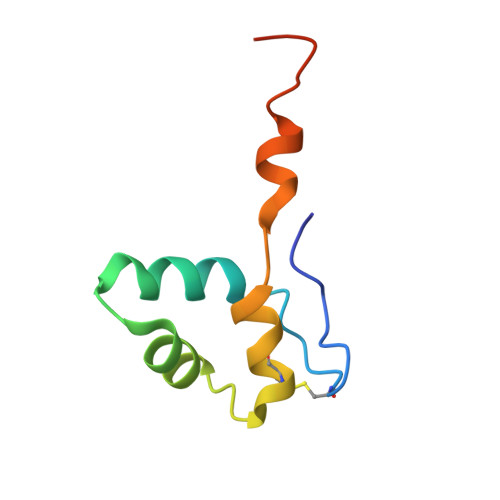
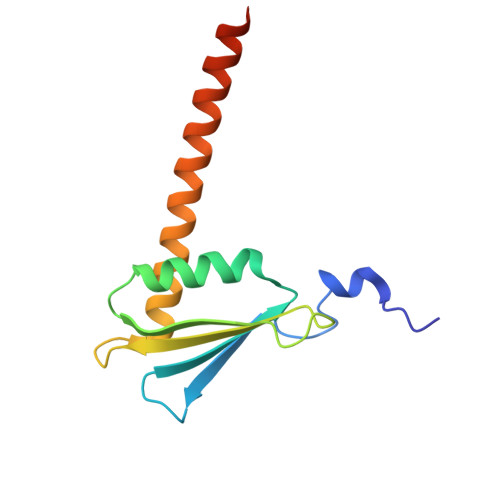
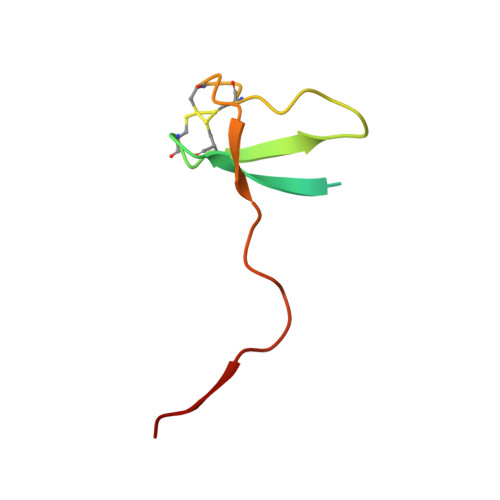
DSIF factor Spt5 coordinates transcription, maturation and exoribonucleolysis of RNA polymerase II transcripts.
Kus, K., Carrique, L., Kecman, T., Fournier, M., Hassanein, S.S., Aydin, E., Kilchert, C., Grimes, J.M., Vasiljeva, L.(2025) Nat Commun 16: 10-10
- PubMed: 39746995
- DOI: https://doi.org/10.1038/s41467-024-55063-7
- Primary Citation of Related Structures:
8QSZ - PubMed Abstract:
Precursor messenger RNA (pre-mRNA) is processed into its functional form during RNA polymerase II (Pol II) transcription. Although functional coupling between transcription and pre-mRNA processing is established, the underlying mechanisms are not fully understood. We show that the key transcription termination factor, RNA exonuclease Xrn2 engages with Pol II forming a stable complex. Xrn2 activity is stimulated by Spt5 to ensure efficient degradation of nascent RNA leading to Pol II dislodgement from DNA. Our results support a model where Xrn2 first forms a stable complex with the elongating Pol II to achieve its full activity in degrading nascent RNA revising the current 'torpedo' model of termination, which posits that RNA degradation precedes Xrn2 engagement with Pol II. Spt5 is also a key factor that attenuates the expression of non-coding transcripts, coordinates pre-mRNA splicing and 3'-end processing. Our findings indicate that engagement with the transcribing Pol II is an essential regulatory step modulating the activity of RNA enzymes such as Xrn2, thus advancing our understanding of how RNA maturation is controlled during transcription.
- Department of Biochemistry, University of Oxford, Oxford, United Kingdom. krzysztof.kus@bioch.ox.ac.uk.
Organizational Affiliation: