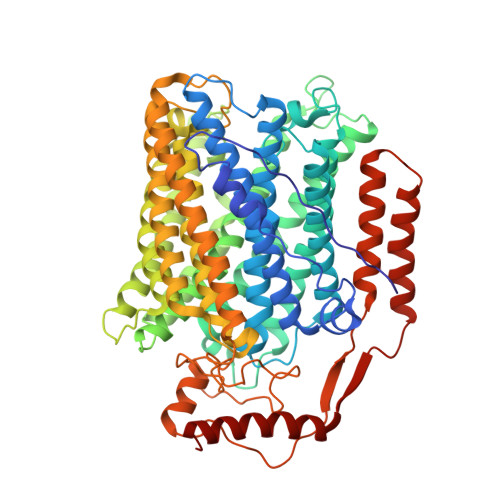
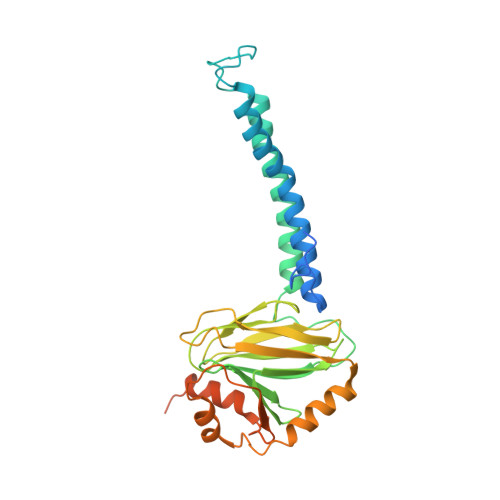
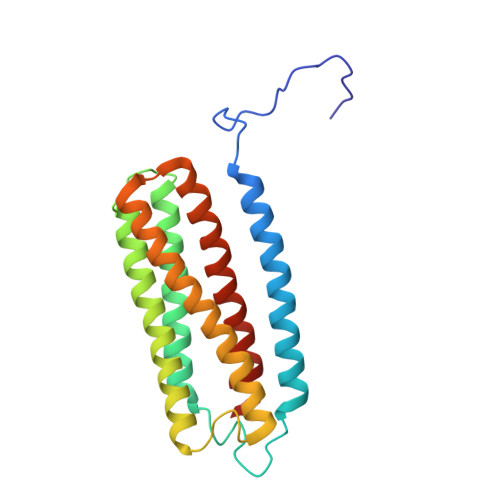
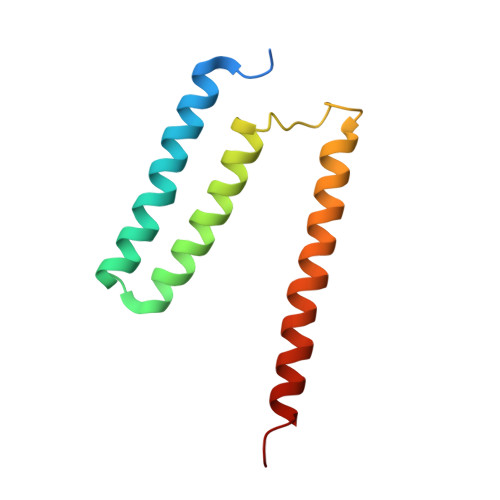
Cryo-EM structure of cytochrome bo 3 quinol oxidase assembled in peptidiscs reveals an "open" conformation for potential ubiquinone-8 release.
Gao, Y., Zhang, Y., Hakke, S., Mohren, R., Sijbers, L.J.P.M., Peters, P.J., Ravelli, R.B.G.(2024) Biochim Biophys Acta Bioenerg 1865: 149045-149045
- PubMed: 38614453
- DOI: https://doi.org/10.1016/j.bbabio.2024.149045
- Primary Citation of Related Structures:
8QQK - PubMed Abstract:
Cytochrome bo 3 quinol oxidase belongs to the heme‑copper-oxidoreductase (HCO) superfamily, which is part of the respiratory chain and essential for cell survival. While the reaction mechanism of cyt bo 3 has been studied extensively over the last decades, specific details about its substrate binding and product release have remained unelucidated due to the lack of structural information. Here, we report a 2.8 Å cryo-electron microscopy structure of cyt bo 3 from Escherichia coli assembled in peptidiscs. Our structural model shows a conformation for amino acids 1-41 of subunit I different from all previously published structures while the remaining parts of this enzyme are similar. Our new conformation shows a "U-shape" assembly in contrast to the transmembrane helix, named "TM0", in other reported structural models. However, TM0 blocks ubiquinone-8 (reaction product) release, suggesting that other cyt bo 3 conformations should exist. Our structural model presents experimental evidence for an "open" conformation to facilitate substrate/product exchange. This work helps further understand the reaction cycle of this oxidase, which could be a benefit for potential drug/antibiotic design for health science.
- Division of Nanoscopy, Maastricht Multimodal Molecular Imaging Institute, Maastricht University, Maastricht, the Netherlands; Division of Imaging Mass Spectrometry, Maastricht Multimodal Molecular Imaging Institute, Maastricht University, Maastricht, the Netherlands. Electronic address: y.gao@maastrichtuniversity.nl.
Organizational Affiliation: