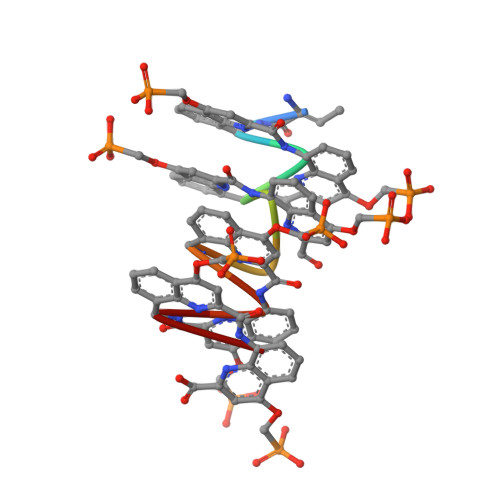
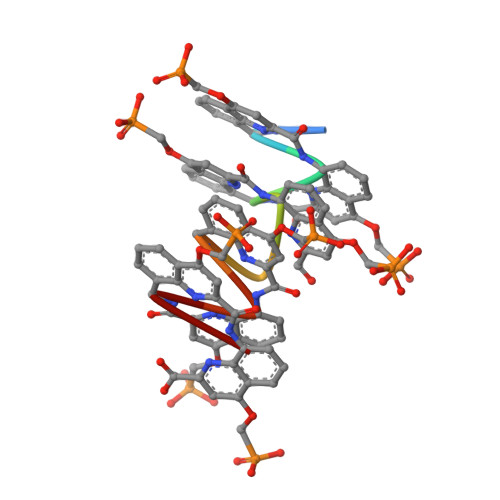
DNA Mimic Foldamer Recognition of a Chromosomal Protein.
Deepak, D., Wu, J., Corvaglia, V., Allmendinger, L., Scheckenbach, M., Tinnefeld, P., Huc, I.(2025) Angew Chem Int Ed Engl 64: e202422958-e202422958
- PubMed: 39714421
- DOI: https://doi.org/10.1002/anie.202422958
- Primary Citation of Related Structures:
8CMN, 8Q2M, 8QPC - PubMed Abstract:
Helical aromatic oligoamide foldamers bearing anionic side chains that mimic the overall shape and charge surface distribution of DNA were synthesized. Their interactions with chromosomal protein Sac7d, a non-sequence-selective DNA-binder that kinks DNA, were investigated by Surface Plasmon Resonance (SPR), Isothermal Titration Calorimetry (ITC), Circular Dichroism spectroscopy (CD), melting curve analysis, Atomic Force Microscopy (AFM), and Nuclear Magnetic Resonance (NMR), as well as by single crystal X-ray crystallography. The foldamers were shown to bind to Sac7d better than a DNA duplex of comparable length. The interaction is diastereoselective and takes place at the DNA binding site. Crystallography revealed that the DNA mimic foldamers have a binding mode of their own and that they can bind to Sac7d without being kinked.
- Department of Pharmacy, Ludwig-Maximilians-Universität München, Butenandtstr. 5-13, 81377, München, Germany.
Organizational Affiliation: