Structure of a nucleosome composed of a palindromic 167-base pair blunt-ended DNA fragment
Ma, Z., Davey, C.A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H3.1 | A [auth AAA], E [auth EEE], K [auth KKK], O [auth OOO] | 98 | Homo sapiens | Mutation(s): 0 Gene Names: | 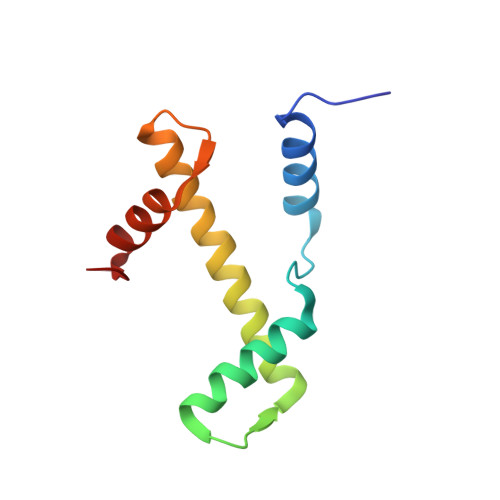 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P68431 (Homo sapiens) Explore P68431 Go to UniProtKB: P68431 | |||||
PHAROS: P68431 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P68431 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H4 | B [auth BBB], F [auth FFF], L [auth LLL], P [auth PPP] | 79 | Homo sapiens | Mutation(s): 0 | 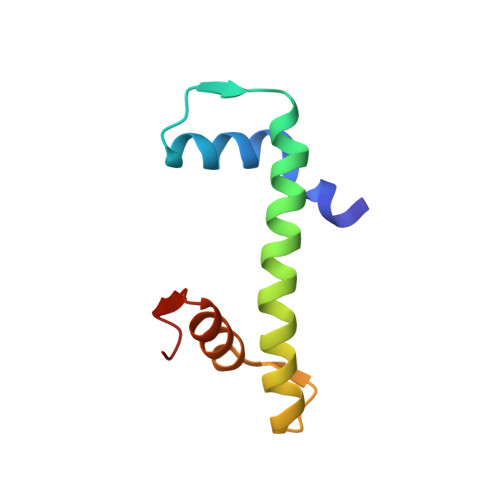 |
UniProt | |||||
Find proteins for A0A8C1JQV0 (Cyprinus carpio) Explore A0A8C1JQV0 Go to UniProtKB: A0A8C1JQV0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A8C1JQV0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2A | C [auth CCC], M [auth MMM] | 105 | Homo sapiens | Mutation(s): 0 Gene Names: H2AC8 | 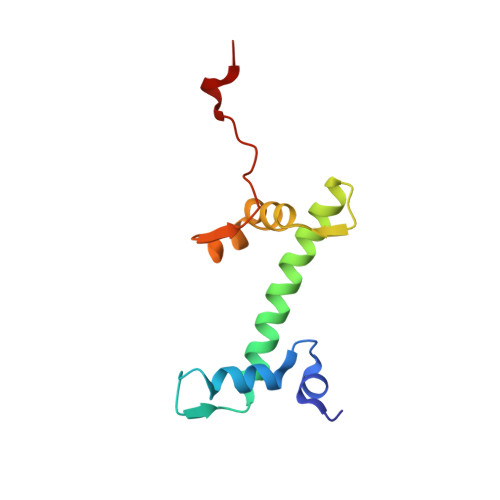 |
UniProt | |||||
Find proteins for H2QSF5 (Pan troglodytes) Explore H2QSF5 Go to UniProtKB: H2QSF5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | H2QSF5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2B type 1-J | D [auth DDD], N [auth NNN] | 96 | Homo sapiens | Mutation(s): 0 Gene Names: H2BC11 | 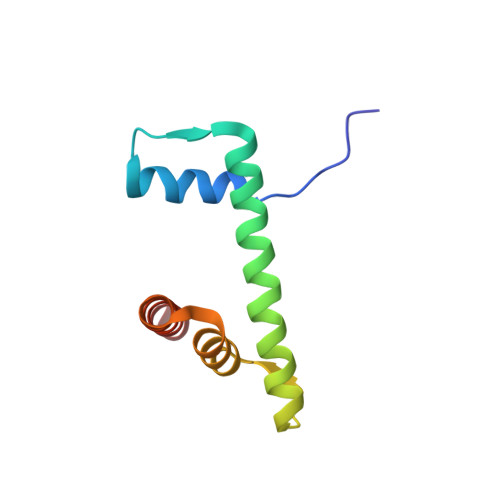 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P06899 (Homo sapiens) Explore P06899 Go to UniProtKB: P06899 | |||||
PHAROS: P06899 GTEx: ENSG00000124635 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P06899 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2A | G [auth GGG], Q [auth QQQ] | 103 | Homo sapiens | Mutation(s): 0 Gene Names: LOC102977259 |  |
UniProt | |||||
Find proteins for A0A2Y9FT95 (Physeter macrocephalus) Explore A0A2Y9FT95 Go to UniProtKB: A0A2Y9FT95 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A2Y9FT95 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Histone H2B type 1-J | H [auth HHH], R [auth RRR] | 97 | Homo sapiens | Mutation(s): 0 Gene Names: H2BC11 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P06899 (Homo sapiens) Explore P06899 Go to UniProtKB: P06899 | |||||
PHAROS: P06899 GTEx: ENSG00000124635 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P06899 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (167-MER) | I [auth III], S [auth SSS] | 167 | synthetic construct |  | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (167-MER) | J [auth JJJ], T [auth TTT] | 167 | synthetic construct |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MN Query on MN | AA [auth III] AB [auth TTT] BA [auth III] CA [auth III] DA [auth III] | MANGANESE (II) ION Mn WAEMQWOKJMHJLA-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 108.437 | α = 90 |
| b = 103.722 | β = 93.59 |
| c = 185.405 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| SCALA | data scaling |
| MOLREP | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Ministry of Education (MoE, Singapore) | Singapore | -- |