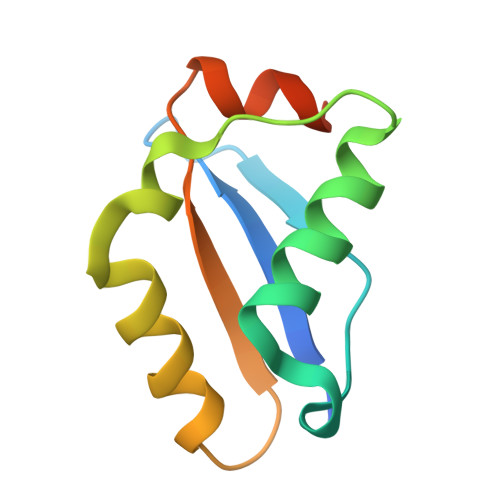
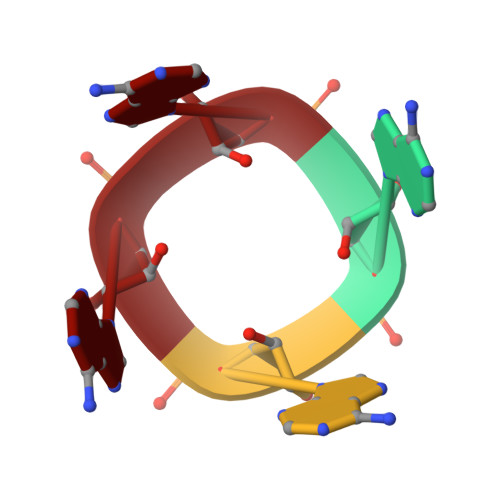
CRISPR antiphage defence mediated by the cyclic nucleotide-binding membrane protein Csx23.
Gruschow, S., McQuarrie, S., Ackermann, K., McMahon, S., Bode, B.E., Gloster, T.M., White, M.F.(2024) Nucleic Acids Res 52: 2761-2775
- PubMed: 38471818
- DOI: https://doi.org/10.1093/nar/gkae167
- Primary Citation of Related Structures:
8QJK - PubMed Abstract:
CRISPR-Cas provides adaptive immunity in prokaryotes. Type III CRISPR systems detect invading RNA and activate the catalytic Cas10 subunit, which generates a range of nucleotide second messengers to signal infection. These molecules bind and activate a diverse range of effector proteins that provide immunity by degrading viral components and/or by disturbing key aspects of cellular metabolism to slow down viral replication. Here, we focus on the uncharacterised effector Csx23, which is widespread in Vibrio cholerae. Csx23 provides immunity against plasmids and phage when expressed in Escherichia coli along with its cognate type III CRISPR system. The Csx23 protein localises in the membrane using an N-terminal transmembrane α-helical domain and has a cytoplasmic C-terminal domain that binds cyclic tetra-adenylate (cA4), activating its defence function. Structural studies reveal a tetrameric structure with a novel fold that binds cA4 specifically. Using pulse EPR, we demonstrate that cA4 binding to the cytoplasmic domain of Csx23 results in a major perturbation of the transmembrane domain, consistent with the opening of a pore and/or disruption of membrane integrity. This work reveals a new class of cyclic nucleotide binding protein and provides key mechanistic detail on a membrane-associated CRISPR effector.
- Biomedical Sciences Research Complex, School of Biology, University of St Andrews, St Andrews, Fife KY16 9ST, UK.
Organizational Affiliation: