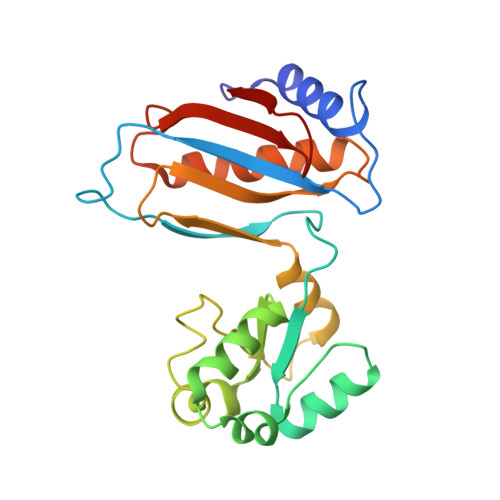
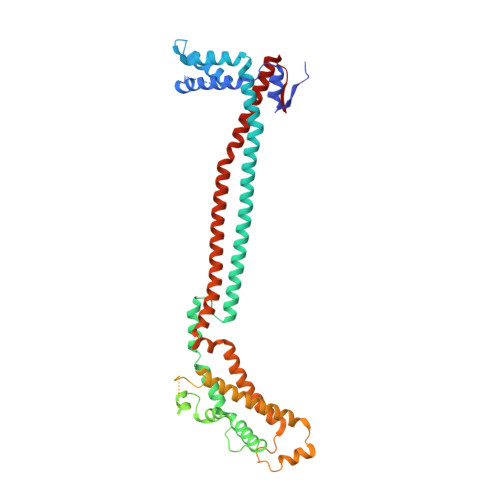
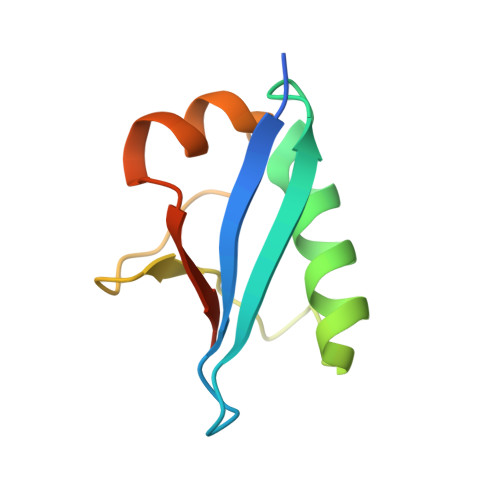
The UFM1 E3 ligase recognizes and releases 60S ribosomes from ER translocons.
Makhlouf, L., Peter, J.J., Magnussen, H.M., Thakur, R., Millrine, D., Minshull, T.C., Harrison, G., Varghese, J., Lamoliatte, F., Foglizzo, M., Macartney, T., Calabrese, A.N., Zeqiraj, E., Kulathu, Y.(2024) Nature 627: 437-444
- PubMed: 38383789
- DOI: https://doi.org/10.1038/s41586-024-07093-w
- Primary Citation of Related Structures:
8BZR, 8C0D, 8QFC, 8QFD - PubMed Abstract:
Stalled ribosomes at the endoplasmic reticulum (ER) are covalently modified with the ubiquitin-like protein UFM1 on the 60S ribosomal subunit protein RPL26 (also known as uL24) 1,2 . This modification, which is known as UFMylation, is orchestrated by the UFM1 ribosome E3 ligase (UREL) complex, comprising UFL1, UFBP1 and CDK5RAP3 (ref. 3 ). However, the catalytic mechanism of UREL and the functional consequences of UFMylation are unclear. Here we present cryo-electron microscopy structures of UREL bound to 60S ribosomes, revealing the basis of its substrate specificity. UREL wraps around the 60S subunit to form a C-shaped clamp architecture that blocks the tRNA-binding sites at one end, and the peptide exit tunnel at the other. A UFL1 loop inserts into and remodels the peptidyl transferase centre. These features of UREL suggest a crucial function for UFMylation in the release and recycling of stalled or terminated ribosomes from the ER membrane. In the absence of functional UREL, 60S-SEC61 translocon complexes accumulate at the ER membrane, demonstrating that UFMylation is necessary for releasing SEC61 from 60S subunits. Notably, this release is facilitated by a functional switch of UREL from a 'writer' to a 'reader' module that recognizes its product-UFMylated 60S ribosomes. Collectively, we identify a fundamental role for UREL in dissociating 60S subunits from the SEC61 translocon and the basis for UFMylation in regulating protein homeostasis at the ER.
- Astbury Centre for Structural Molecular Biology, School of Molecular and Cellular Biology, Faculty of Biological Sciences, University of Leeds, Leeds, UK.
Organizational Affiliation: