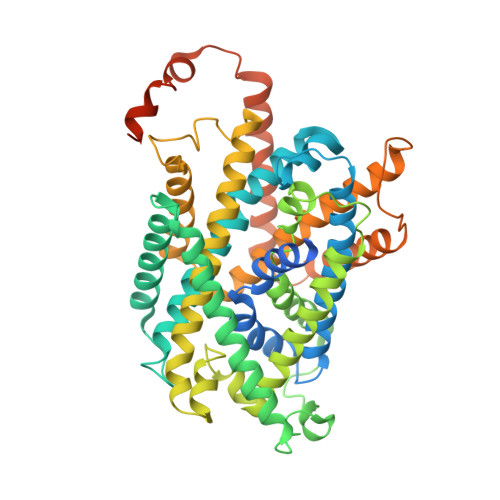
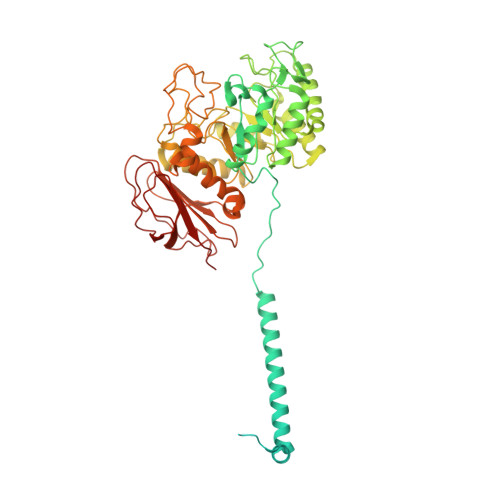
Structure and mechanisms of transport of human Asc1/CD98hc amino acid transporter.
Rullo-Tubau, J., Martinez-Molledo, M., Bartoccioni, P., Puch-Giner, I., Arias, A., Saen-Oon, S., Stephan-Otto Attolini, C., Artuch, R., Diaz, L., Guallar, V., Errasti-Murugarren, E., Palacin, M., Llorca, O.(2024) Nat Commun 15: 2986-2986
- PubMed: 38582862
- DOI: https://doi.org/10.1038/s41467-024-47385-3
- Primary Citation of Related Structures:
8QEY - PubMed Abstract:
Recent cryoEM studies elucidated details of the structural basis for the substrate selectivity and translocation of heteromeric amino acid transporters. However, Asc1/CD98hc is the only neutral heteromeric amino acid transporter that can function through facilitated diffusion, and the only one that efficiently transports glycine and D-serine, and thus has a regulatory role in the central nervous system. Here we use cryoEM, ligand-binding simulations, mutagenesis, transport assays, and molecular dynamics to define human Asc1/CD98hc determinants for substrate specificity and gain insights into the mechanisms that govern substrate translocation by exchange and facilitated diffusion. The cryoEM structure of Asc1/CD98hc is determined at 3.4-3.8 Å resolution, revealing an inward-facing semi-occluded conformation. We find that Ser 246 and Tyr 333 are essential for Asc1/CD98hc substrate selectivity and for the exchange and facilitated diffusion modes of transport. Taken together, these results reveal the structural bases for ligand binding and transport features specific to human Asc1.
- Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology (BIST), Baldiri Reixac 10, E-08028, Barcelona, Spain.
Organizational Affiliation: