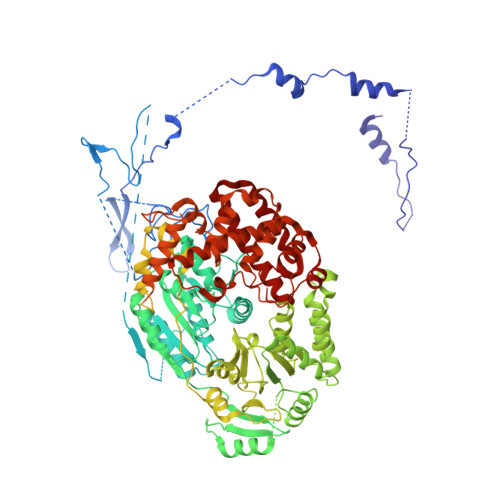
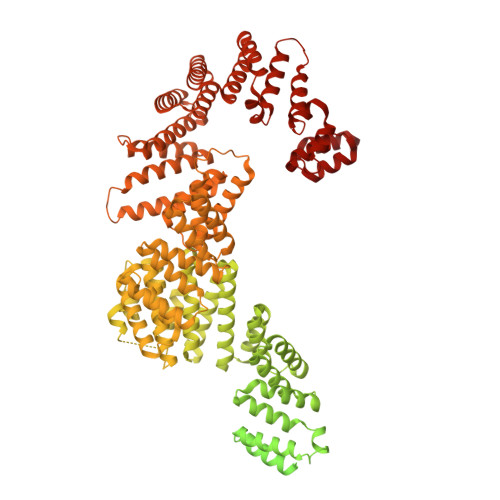
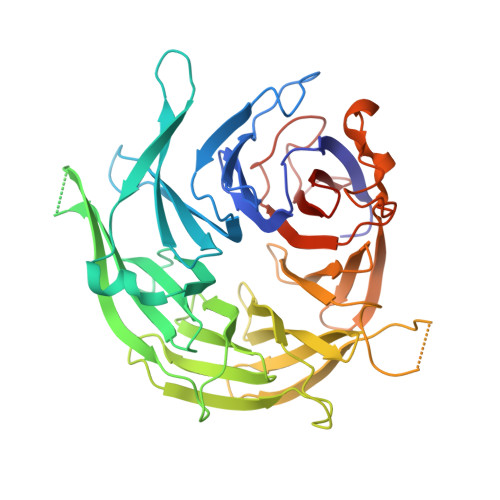
Concerted structural rearrangements enable RNA channeling into the cytoplasmic Ski238-Ski7-exosome assembly.
Keidel, A., Kogel, A., Reichelt, P., Kowalinski, E., Schafer, I.B., Conti, E.(2023) Mol Cell 83: 4093
- PubMed: 37879335
- DOI: https://doi.org/10.1016/j.molcel.2023.09.037
- Primary Citation of Related Structures:
8Q9T, 8QCA, 8QCB, 8QCF - PubMed Abstract:
The Ski2-Ski3-Ski8 (Ski238) helicase complex directs cytoplasmic mRNAs toward the nucleolytic exosome complex for degradation. In yeast, the interaction between Ski238 and exosome requires the adaptor protein Ski7. We determined different cryo-EM structures of the Ski238 complex depicting the transition from a rigid autoinhibited closed conformation to a flexible active open conformation in which the Ski2 helicase module has detached from the rest of Ski238. The open conformation favors the interaction of the Ski3 subunit with exosome-bound Ski7, leading to the recruitment of the exosome. In the Ski238-Ski7-exosome holocomplex, the Ski2 helicase module binds the exosome cap, enabling the RNA to traverse from the helicase through the internal exosome channel to the Rrp44 exoribonuclease. Our study pinpoints how conformational changes within the Ski238 complex regulate exosome recruitment for RNA degradation. We also reveal the remarkable conservation of helicase-exosome RNA channeling mechanisms throughout eukaryotic nuclear and cytoplasmic exosome complexes.
- Department of Structural Cell Biology, Max Planck Institute of Biochemistry, Am Klopferspitz 18, Martinsried, 82152 Munich, Germany.
Organizational Affiliation: