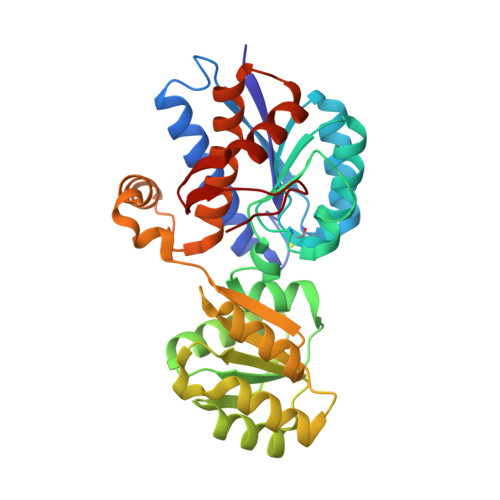
Molecular handcraft of a well-folded protein chimera.
Toledo-Patino, S., Goetz, S.K., Shanmugaratnam, S., Hocker, B., Farias-Rico, J.A.(2024) FEBS Lett 598: 1375-1386
- PubMed: 38508768
- DOI: https://doi.org/10.1002/1873-3468.14856
- Primary Citation of Related Structures:
8Q52 - PubMed Abstract:
Modular assembly is a compelling pathway to create new proteins, a concept supported by protein engineering and millennia of evolution. Natural evolution provided a repository of building blocks, known as domains, which trace back to even shorter segments that underwent numerous 'copy-paste' processes culminating in the scaffolds we see today. Utilizing the subdomain-database Fuzzle, we constructed a fold-chimera by integrating a flavodoxin-like fragment into a periplasmic binding protein. This chimera is well-folded and a crystal structure reveals stable interfaces between the fragments. These findings demonstrate the adaptability of α/β-proteins and offer a stepping stone for optimization. By emphasizing the practicality of fragment databases, our work pioneers new pathways in protein engineering. Ultimately, the results substantiate the conjecture that periplasmic binding proteins originated from a flavodoxin-like ancestor.
- Max Planck Institute for Developmental Biology, Tübingen, Germany.
Organizational Affiliation: