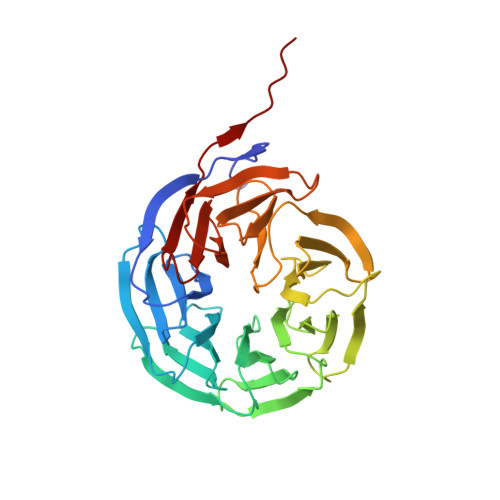
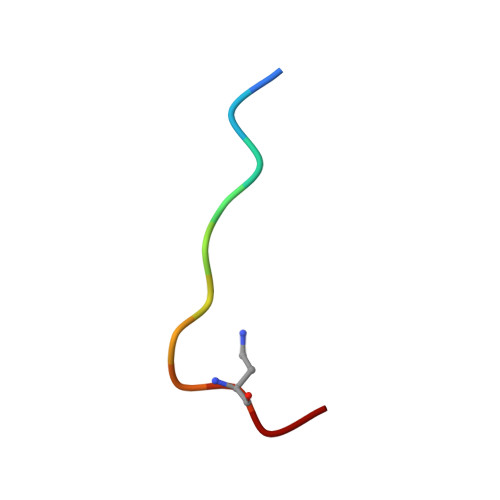
Macrocyclic peptides as inhibitors of WDR5-lncRNA interactions.
Chang, J.Y., Neugebauer, C., Schmeing, S., Amrahova, G., 't Hart, P.(2023) Chem Commun (Camb) 59: 10656-10659
- PubMed: 37581220
- DOI: https://doi.org/10.1039/d3cc03221c
- Primary Citation of Related Structures:
8Q1N - PubMed Abstract:
WDR5 is an adaptor protein involved in the regulation of various epigenetic modifier complexes. Various inhibitors have been described but only as inhibitors of its protein-protein interactions. Here we describe peptidic macrocycles that act as inhibitors of the interaction between WDR5 and long non-coding RNAs. The findings provide a new strategy to modulate the biological function of WDR5 as an RNA binding epigenetic regulator.
- Chemical Genomics Centre of the Max Planck, Society Max Planck Institute of Molecular Physiology, Otto-Hahn-Strasse 11, 44227 Dortmund, Germany. peter.t-hart@mpi-dortmund.mpg.de.
Organizational Affiliation: