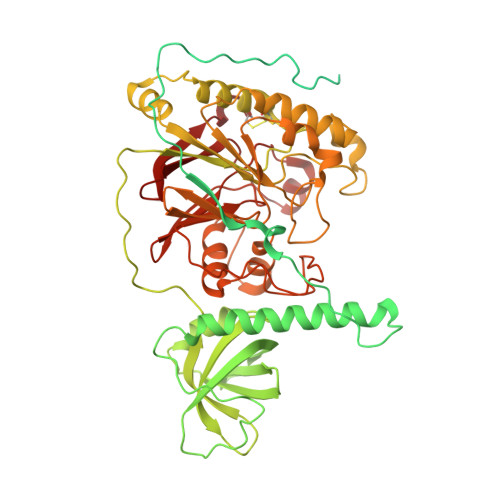
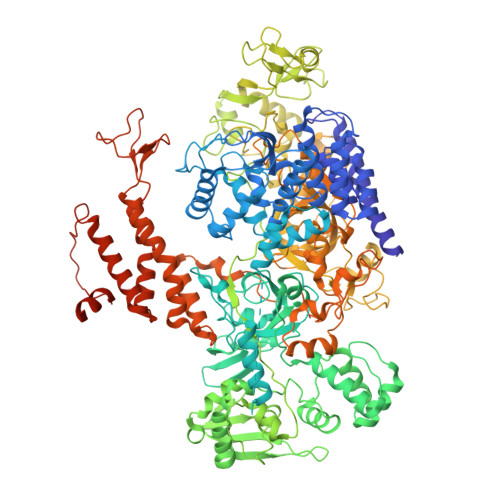
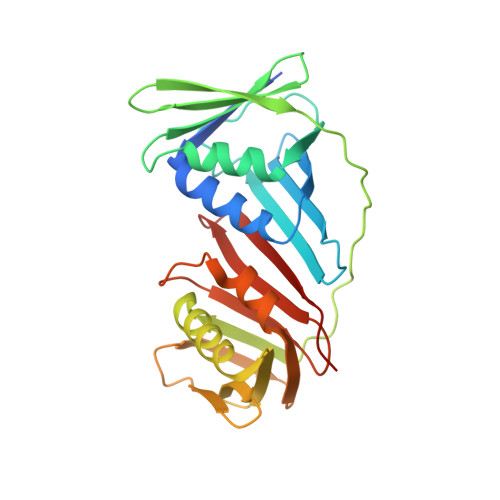
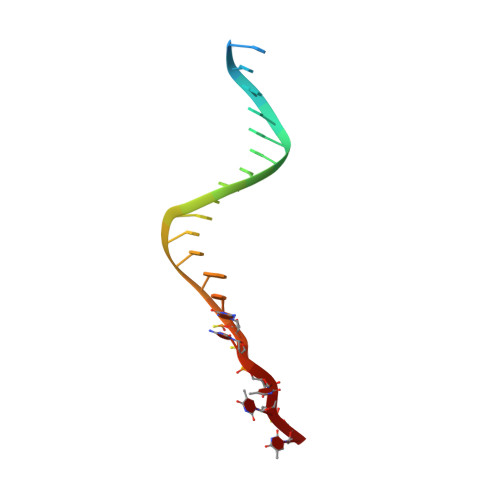
Molecular basis for proofreading by the unique exonuclease domain of Family-D DNA polymerases.
Betancurt-Anzola, L., Martinez-Carranza, M., Delarue, M., Zatopek, K.M., Gardner, A.F., Sauguet, L.(2023) Nat Commun 14: 8306-8306
- PubMed: 38097591
- DOI: https://doi.org/10.1038/s41467-023-44125-x
- Primary Citation of Related Structures:
8PPT, 8PPU, 8PPV - PubMed Abstract:
Replicative DNA polymerases duplicate entire genomes at high fidelity. This feature is shared among the three domains of life and is facilitated by their dual polymerase and exonuclease activities. Family D replicative DNA polymerases (PolD), found exclusively in Archaea, contain an unusual RNA polymerase-like catalytic core, and a unique Mre11-like proofreading active site. Here, we present cryo-EM structures of PolD trapped in a proofreading mode, revealing an unanticipated correction mechanism that extends the repertoire of protein domains known to be involved in DNA proofreading. Based on our experimental structures, mutants of PolD were designed and their contribution to mismatch bypass and exonuclease kinetics was determined. This study sheds light on the convergent evolution of structurally distinct families of DNA polymerases, and the domain acquisition and exchange mechanism that occurred during the evolution of the replisome in the three domains of life.
- Architecture and Dynamics of Biological Macromolecules, Institut Pasteur, Université Paris Cité, CNRS, UMR 3528, Paris, France.
Organizational Affiliation: