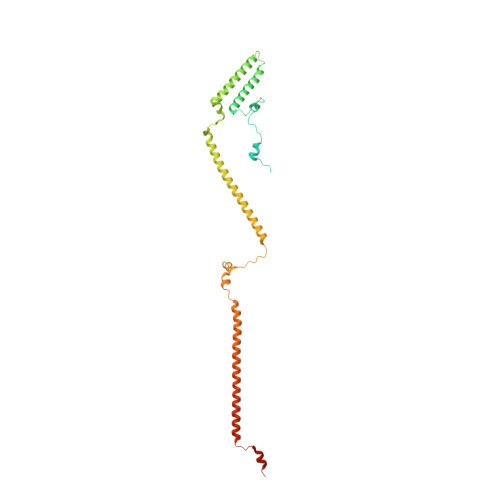
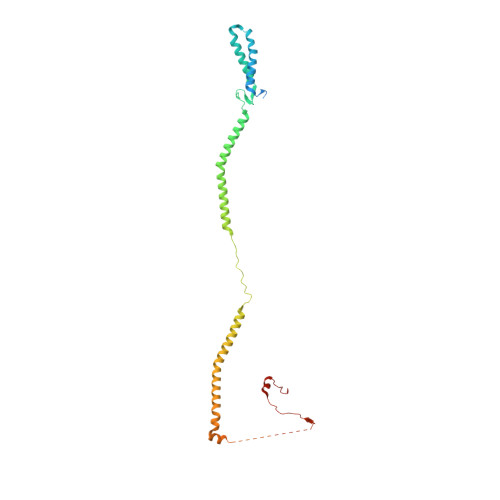
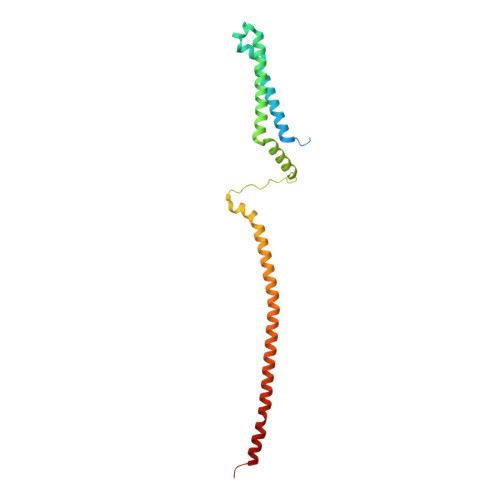
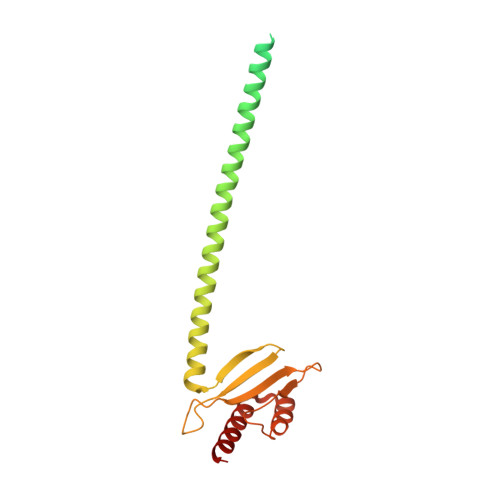
Structure of the human outer kinetochore KMN network complex.
Yatskevich, S., Yang, J., Bellini, D., Zhang, Z., Barford, D.(2024) Nat Struct Mol Biol 31: 874-883
- PubMed: 38459127
- DOI: https://doi.org/10.1038/s41594-024-01249-y
- Primary Citation of Related Structures:
8PPR - PubMed Abstract:
Faithful chromosome segregation requires robust, load-bearing attachments of chromosomes to the mitotic spindle, a function accomplished by large macromolecular complexes termed kinetochores. In most eukaryotes, the constitutive centromere-associated network (CCAN) complex of the inner kinetochore recruits to centromeres the ten-subunit outer kinetochore KMN network that comprises the KNL1C, MIS12C and NDC80C complexes. The KMN network directly attaches CCAN to microtubules through MIS12C and NDC80C. Here, we determined a high-resolution cryo-EM structure of the human KMN network. This showed an intricate and extensive assembly of KMN subunits, with the central MIS12C forming rigid interfaces with NDC80C and KNL1C, augmented by multiple peptidic inter-subunit connections. We also observed that unphosphorylated MIS12C exists in an auto-inhibited state that suppresses its capacity to interact with CCAN. Ser100 and Ser109 of the N-terminal segment of the MIS12C subunit Dsn1, two key targets of Aurora B kinase, directly stabilize this auto-inhibition. Our study indicates how selectively relieving this auto-inhibition through Ser100 and Ser109 phosphorylation might restrict outer kinetochore assembly to functional centromeres during cell division.
- MRC Laboratory of Molecular Biology, Francis Crick Avenue, Cambridge, UK. yatskevs@gene.com.
Organizational Affiliation: