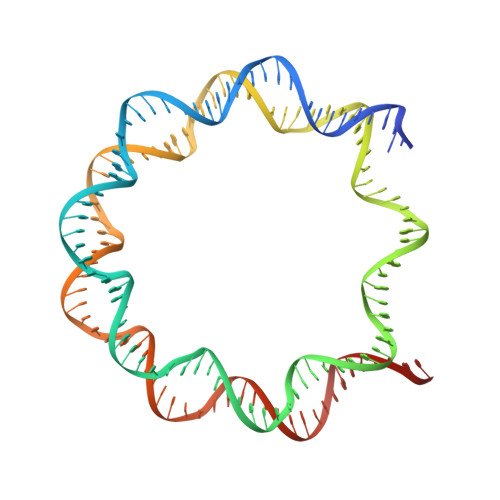
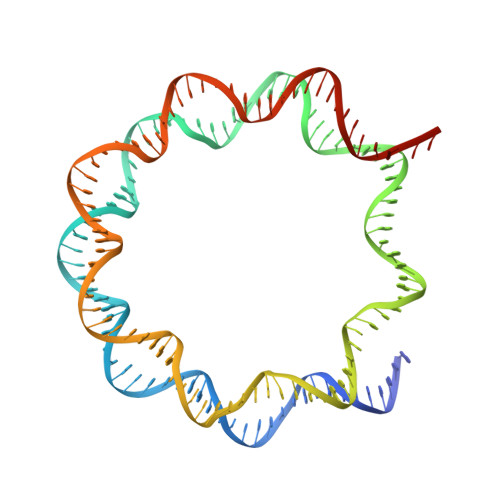
Nucleosome-bound NR5A2 structure reveals pioneer factor mechanism by DNA minor groove anchor competition.
Kobayashi, W., Sappler, A.H., Bollschweiler, D., Kummecke, M., Basquin, J., Arslantas, E.N., Ruangroengkulrith, S., Hornberger, R., Duderstadt, K., Tachibana, K.(2024) Nat Struct Mol Biol 31: 757-766
- PubMed: 38409506
- DOI: https://doi.org/10.1038/s41594-024-01239-0
- Primary Citation of Related Structures:
8PKI, 8PKJ - PubMed Abstract:
Gene expression during natural and induced reprogramming is controlled by pioneer transcription factors that initiate transcription from closed chromatin. Nr5a2 is a key pioneer factor that regulates zygotic genome activation in totipotent embryos, pluripotency in embryonic stem cells and metabolism in adult tissues, but the mechanism of its pioneer activity remains poorly understood. Here, we present a cryo-electron microscopy structure of human NR5A2 bound to a nucleosome. The structure shows that the conserved carboxy-terminal extension (CTE) loop of the NR5A2 DNA-binding domain competes with a DNA minor groove anchor of the nucleosome and releases entry-exit site DNA. Mutational analysis showed that NR5A2 D159 of the CTE is dispensable for DNA binding but required for stable nucleosome association and persistent DNA 'unwrapping'. These findings suggest that NR5A2 belongs to an emerging class of pioneer factors that can use DNA minor groove anchor competition to destabilize nucleosomes and facilitate gene expression during reprogramming.
- Department of Totipotency, Max Planck Institute of Biochemistry (MPIB), Munich, Germany.
Organizational Affiliation: