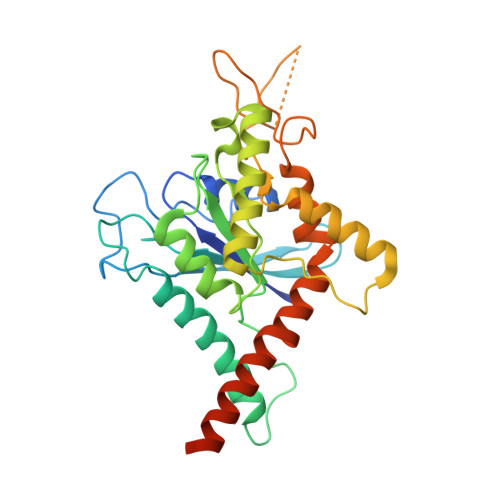
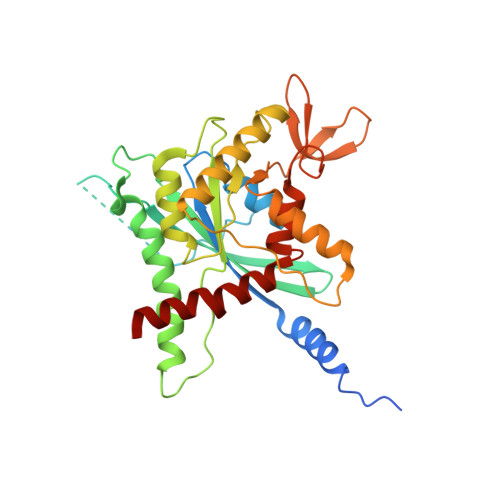
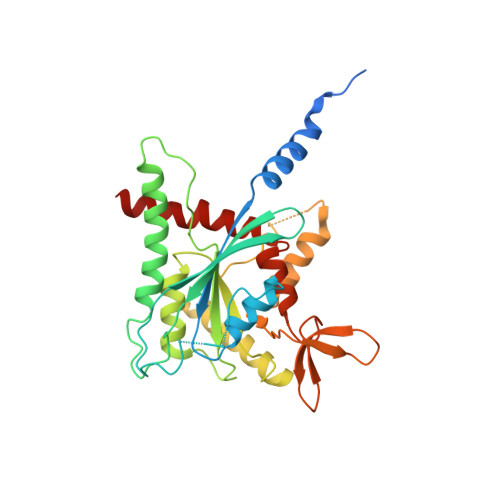
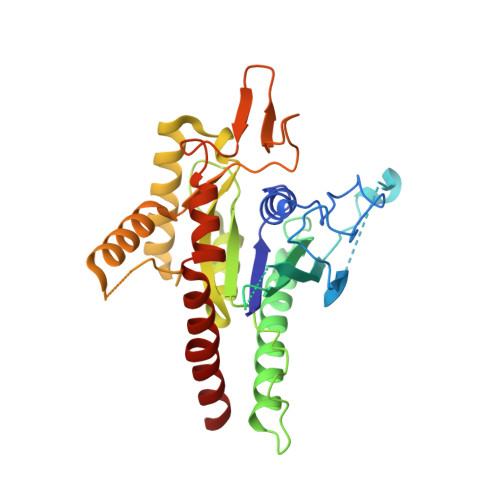
The structure of a tetrameric septin complex reveals a hydrophobic element essential for NC-interface integrity.
Grupp, B., Denkhaus, L., Gerhardt, S., Vogele, M., Johnsson, N., Gronemeyer, T.(2024) Commun Biol 7: 48-48
- PubMed: 38184752
- DOI: https://doi.org/10.1038/s42003-023-05734-w
- Primary Citation of Related Structures:
8PFH - PubMed Abstract:
The septins of the yeast Saccharomyces cerevisiae assemble into hetero-octameric rods by alternating interactions between neighboring G-domains or N- and C-termini, respectively. These rods polymerize end to end into apolar filaments, forming a ring beneath the prospective new bud that expands during the cell cycle into an hourglass structure. The hourglass finally splits during cytokinesis into a double ring. Understanding these transitions as well as the plasticity of the higher order assemblies requires a detailed knowledge of the underlying structures. Here we present the first X-ray crystal structure of a tetrameric Shs1-Cdc12-Cdc3-Cdc10 complex at a resolution of 3.2 Å. Close inspection of the NC-interfaces of this and other septin structures reveals a conserved contact motif that is essential for NC-interface integrity of yeast and human septins in vivo and in vitro. Using the tetrameric structure in combination with AlphaFold-Multimer allowed us to propose a model of the octameric septin rod.
- Institute of Molecular Genetics and Cell Biology, Ulm University, Ulm, Germany.
Organizational Affiliation: