Subunit abundance shapes GABAA receptor stoichiometry
Sente, A., Malinauskas, T., Naydenova, K., Hardwick, S.W., Chirgadze, D.Y., Aricescu, A.R.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Gamma-aminobutyric acid receptor subunit alpha-1 | A, D [auth C] | 484 | Homo sapiens | Mutation(s): 0 Gene Names: GABRA1 | 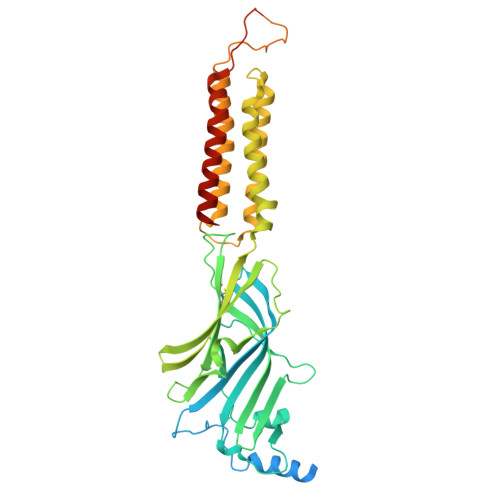 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P14867 (Homo sapiens) Explore P14867 Go to UniProtKB: P14867 | |||||
PHAROS: P14867 GTEx: ENSG00000022355 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P14867 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | Go to GlyGen: P14867-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Gamma-aminobutyric acid receptor subunit beta-3 | B, C [auth D], E | 490 | Homo sapiens | Mutation(s): 0 Gene Names: GABRB3 | 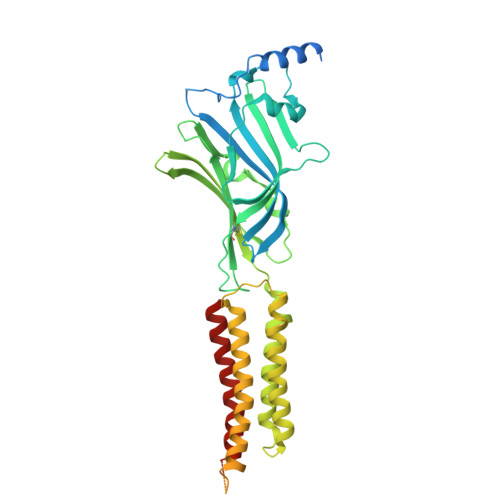 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P28472 (Homo sapiens) Explore P28472 Go to UniProtKB: P28472 | |||||
PHAROS: P28472 GTEx: ENSG00000166206 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P28472 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | Go to GlyGen: P28472-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nanobody Nb25 | 135 | Lama glama | Mutation(s): 0 | 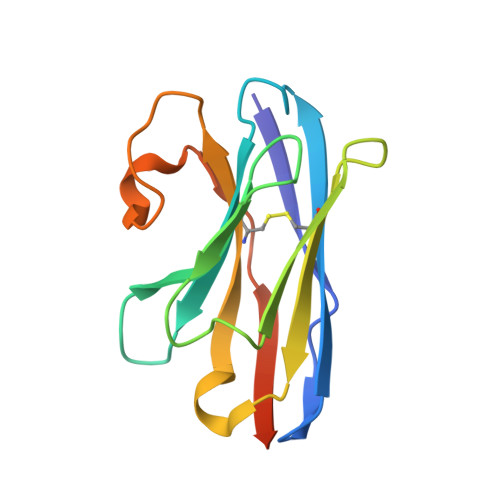 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | G [auth a], H [auth b], J [auth c], L [auth d], M [auth e] | 5 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G22768VO GlyCosmos: G22768VO GlyGen: G22768VO | |||||
| Ligands 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PIO Query on PIO | BA [auth C], P [auth A] | [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate C25 H49 O19 P3 XLNCEHRXXWQMPK-MJUMVPIBSA-N |  | ||
| R16 Query on R16 | AA [auth C], O [auth A] | HEXADECANE C16 H34 DCAYPVUWAIABOU-UHFFFAOYSA-N |  | ||
| D10 Query on D10 | CA [auth E] DA [auth E] EA [auth E] Q [auth B] R [auth B] | DECANE C10 H22 DIOQZVSQGTUSAI-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | U [auth B] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| MODEL REFINEMENT | PHENIX | |
| RECONSTRUCTION | cryoSPARC | 4.1.2 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MR/L009609/1 |
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MC_UP_1201/15 |
| National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) | United States | 1R01GM135550-01 |
| Medical Research Council (MRC, United Kingdom) | United Kingdom | MC_EX_MR/T046279/1 |
| National Science Foundation (NSF, United States) | United States | 2014862 |