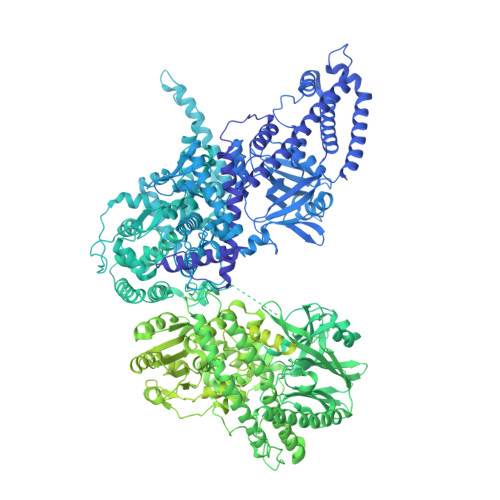
Structure and activation mechanism of the Makes caterpillars floppy 1 toxin.
Belyy, A., Heilen, P., Hagel, P., Hofnagel, O., Raunser, S.(2023) Nat Commun 14: 8226-8226
- PubMed: 38086871
- DOI: https://doi.org/10.1038/s41467-023-44069-2
- Primary Citation of Related Structures:
8P50, 8P51, 8P52 - PubMed Abstract:
The bacterial Makes caterpillars floppy 1 (Mcf1) toxin promotes apoptosis in insects, leading to loss of body turgor and death. The molecular mechanism underlying Mcf1 intoxication is poorly understood. Here, we present the cryo-EM structure of Mcf1 from Photorhabdus luminescens, revealing a seahorse-like shape with a head and tail. While the three head domains contain two effectors, as well as an activator-binding domain (ABD) and an autoprotease, the tail consists of two putative translocation and three putative receptor-binding domains. Rearrangement of the tail moves the C-terminus away from the ABD and allows binding of the host cell ADP-ribosylation factor 3, inducing conformational changes that position the cleavage site closer to the protease. This distinct activation mechanism that is based on a hook-loop interaction results in three autocleavage reactions and the release of two toxic effectors. Unexpectedly, the BH3-like domain containing ABD is not an active effector. Our findings allow us to understand key steps of Mcf1 intoxication at the molecular level.
- Department of Structural Biochemistry, Max Planck Institute of Molecular Physiology, Otto-Hahn-Str. 11, 44227, Dortmund, Germany.
Organizational Affiliation: