The compensatory mechanism of a naturally evolved E167K RF2 counteracting the loss of RF1 in bacteria
Pundir, S., Larsson, D.S.D., Selmer, M., Sanyal, S.To be published.
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS2 | B [auth g] | 241 | Escherichia coli | Mutation(s): 0 | 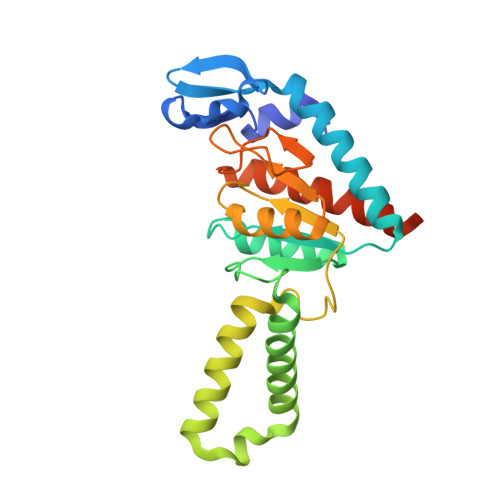 |
UniProt | |||||
Find proteins for P0A7V0 (Escherichia coli (strain K12)) Explore P0A7V0 Go to UniProtKB: P0A7V0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7V0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS3 | C [auth h] | 233 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7V3 (Escherichia coli (strain K12)) Explore P0A7V3 Go to UniProtKB: P0A7V3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7V3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS4 | D [auth i] | 206 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7V8 (Escherichia coli (strain K12)) Explore P0A7V8 Go to UniProtKB: P0A7V8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7V8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS5 | E [auth j] | 167 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7W1 (Escherichia coli (strain K12)) Explore P0A7W1 Go to UniProtKB: P0A7W1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7W1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S6 | F [auth k] | 135 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P02358 (Escherichia coli (strain K12)) Explore P02358 Go to UniProtKB: P02358 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02358 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 30S ribosomal protein S7 | G [auth l] | 179 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P02359 (Escherichia coli (strain K12)) Explore P02359 Go to UniProtKB: P02359 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02359 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS8 | H [auth m] | 130 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7W7 (Escherichia coli (strain K12)) Explore P0A7W7 Go to UniProtKB: P0A7W7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7W7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS9 | I [auth n] | 130 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7X3 (Escherichia coli (strain K12)) Explore P0A7X3 Go to UniProtKB: P0A7X3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7X3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS10 | J [auth o] | 103 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7R5 (Escherichia coli (strain K12)) Explore P0A7R5 Go to UniProtKB: P0A7R5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7R5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS11 | K [auth p] | 129 | Escherichia coli | Mutation(s): 1 |  |
UniProt | |||||
Find proteins for P0A7R9 (Escherichia coli (strain K12)) Explore P0A7R9 Go to UniProtKB: P0A7R9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7R9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS12 | L [auth q] | 124 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7S3 (Escherichia coli (strain K12)) Explore P0A7S3 Go to UniProtKB: P0A7S3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7S3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS13 | M [auth r] | 118 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7S9 (Escherichia coli (strain K12)) Explore P0A7S9 Go to UniProtKB: P0A7S9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7S9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 14 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS14 | N [auth s] | 101 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0AG59 (Escherichia coli (strain K12)) Explore P0AG59 Go to UniProtKB: P0AG59 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AG59 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 15 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS15 | O [auth t] | 89 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0ADZ4 (Escherichia coli (strain K12)) Explore P0ADZ4 Go to UniProtKB: P0ADZ4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ADZ4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 16 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein bS16 | P [auth u] | 82 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7T3 (Escherichia coli (strain K12)) Explore P0A7T3 Go to UniProtKB: P0A7T3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7T3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 17 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS17 | Q [auth v] | 84 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0AG63 (Escherichia coli (strain K12)) Explore P0AG63 Go to UniProtKB: P0AG63 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AG63 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 18 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein bS18 | R [auth w] | 75 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7T7 (Escherichia coli (strain K12)) Explore P0A7T7 Go to UniProtKB: P0A7T7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7T7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 19 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein uS19 | S [auth x] | 92 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7U3 (Escherichia coli (strain K12)) Explore P0A7U3 Go to UniProtKB: P0A7U3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7U3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 20 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein bS20 | T [auth y] | 87 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7U7 (Escherichia coli (strain K12)) Explore P0A7U7 Go to UniProtKB: P0A7U7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7U7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 21 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Small ribosomal subunit protein bS21 | U [auth z] | 71 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P68679 (Escherichia coli (strain K12)) Explore P68679 Go to UniProtKB: P68679 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P68679 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 26 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L2 | Z [auth B] | 273 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P60422 (Escherichia coli (strain K12)) Explore P60422 Go to UniProtKB: P60422 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60422 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 27 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L3 | AA [auth C] | 209 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P60438 (Escherichia coli (strain K12)) Explore P60438 Go to UniProtKB: P60438 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60438 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 28 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L4 | BA [auth D] | 201 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P60723 (Escherichia coli (strain K12)) Explore P60723 Go to UniProtKB: P60723 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60723 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 29 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L5 | CA [auth E] | 179 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P62399 (Escherichia coli (strain K12)) Explore P62399 Go to UniProtKB: P62399 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P62399 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 30 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L6 | DA [auth F] | 177 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0AG55 (Escherichia coli (strain K12)) Explore P0AG55 Go to UniProtKB: P0AG55 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AG55 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 31 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L13 | EA [auth J] | 142 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0AA10 (Escherichia coli (strain K12)) Explore P0AA10 Go to UniProtKB: P0AA10 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AA10 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 32 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L14 | FA [auth K] | 123 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0ADY3 (Escherichia coli (strain K12)) Explore P0ADY3 Go to UniProtKB: P0ADY3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ADY3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 33 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L15 | GA [auth L] | 144 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P02413 (Escherichia coli (strain K12)) Explore P02413 Go to UniProtKB: P02413 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P02413 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 34 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Large ribosomal subunit protein uL16 | HA [auth M] | 136 | Escherichia coli | Mutation(s): 1 |  |
UniProt | |||||
Find proteins for P0ADY7 (Escherichia coli (strain K12)) Explore P0ADY7 Go to UniProtKB: P0ADY7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ADY7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 35 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L17 | IA [auth N] | 127 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0AG44 (Escherichia coli (strain K12)) Explore P0AG44 Go to UniProtKB: P0AG44 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AG44 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 36 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L18 | JA [auth O] | 117 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0C018 (Escherichia coli (strain K12)) Explore P0C018 Go to UniProtKB: P0C018 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0C018 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 37 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L19 | KA [auth P] | 115 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7K6 (Escherichia coli (strain K12)) Explore P0A7K6 Go to UniProtKB: P0A7K6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7K6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 38 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L20 | LA [auth Q] | 118 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7L3 (Escherichia coli (strain K12)) Explore P0A7L3 Go to UniProtKB: P0A7L3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7L3 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 39 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L21 | MA [auth R] | 103 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0AG48 (Escherichia coli (strain K12)) Explore P0AG48 Go to UniProtKB: P0AG48 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AG48 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 40 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L22 | NA [auth S] | 110 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P61175 (Escherichia coli (strain K12)) Explore P61175 Go to UniProtKB: P61175 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61175 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 41 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L23 | OA [auth T] | 100 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0ADZ0 (Escherichia coli (strain K12)) Explore P0ADZ0 Go to UniProtKB: P0ADZ0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0ADZ0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 42 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L24 | PA [auth U] | 104 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P60624 (Escherichia coli (strain K12)) Explore P60624 Go to UniProtKB: P60624 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P60624 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 43 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L25 | QA [auth V] | 94 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P68919 (Escherichia coli (strain K12)) Explore P68919 Go to UniProtKB: P68919 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P68919 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 44 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L27 | RA [auth W] | 85 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7L8 (Escherichia coli (strain K12)) Explore P0A7L8 Go to UniProtKB: P0A7L8 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7L8 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 45 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L28 | SA [auth X] | 78 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7M2 (Escherichia coli (strain K12)) Explore P0A7M2 Go to UniProtKB: P0A7M2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7M2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 46 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L29 | TA [auth Y] | 63 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7M6 (Escherichia coli (strain K12)) Explore P0A7M6 Go to UniProtKB: P0A7M6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7M6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 47 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L30 | UA [auth Z] | 59 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0AG51 (Escherichia coli (strain K12)) Explore P0AG51 Go to UniProtKB: P0AG51 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0AG51 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 48 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L32 | VA [auth b] | 57 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7N4 (Escherichia coli (strain K12)) Explore P0A7N4 Go to UniProtKB: P0A7N4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7N4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 49 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L33 | WA [auth c] | 55 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7N9 (Escherichia coli (strain K12)) Explore P0A7N9 Go to UniProtKB: P0A7N9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7N9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 50 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L34 | XA [auth d] | 46 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7P5 (Escherichia coli (strain K12)) Explore P0A7P5 Go to UniProtKB: P0A7P5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7P5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 51 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L35 | YA [auth e] | 65 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7Q1 (Escherichia coli (strain K12)) Explore P0A7Q1 Go to UniProtKB: P0A7Q1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7Q1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 52 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L36 | ZA [auth f] | 38 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7Q6 (Escherichia coli (strain K12)) Explore P0A7Q6 Go to UniProtKB: P0A7Q6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7Q6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 53 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Large ribosomal subunit protein bL31A | AB [auth a] | 70 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7M9 (Escherichia coli (strain K12)) Explore P0A7M9 Go to UniProtKB: P0A7M9 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7M9 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 54 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L11 | BB [auth I] | 142 | Escherichia coli | Mutation(s): 0 |  |
UniProt | |||||
Find proteins for P0A7J7 (Escherichia coli (strain K12)) Explore P0A7J7 Go to UniProtKB: P0A7J7 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7J7 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 55 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Peptide chain release factor RF2 | CB [auth 6] | 365 | Escherichia coli K-12 | Mutation(s): 1 Gene Names: prfB, supK, b2891, JW5847 | 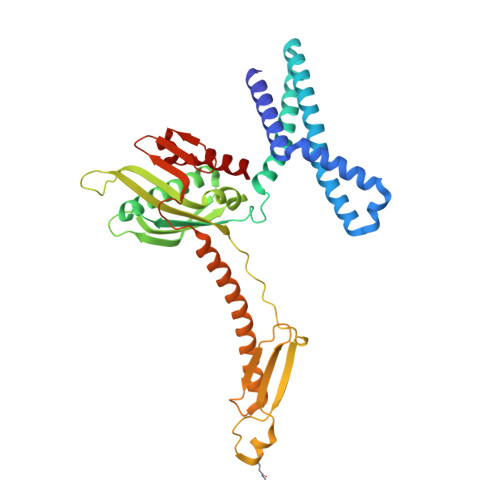 |
UniProt | |||||
Find proteins for P07012 (Escherichia coli (strain K12)) Explore P07012 Go to UniProtKB: P07012 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P07012 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 56 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| 50S ribosomal protein L9 | DB [auth G] | 149 | Escherichia coli | Mutation(s): 0 | 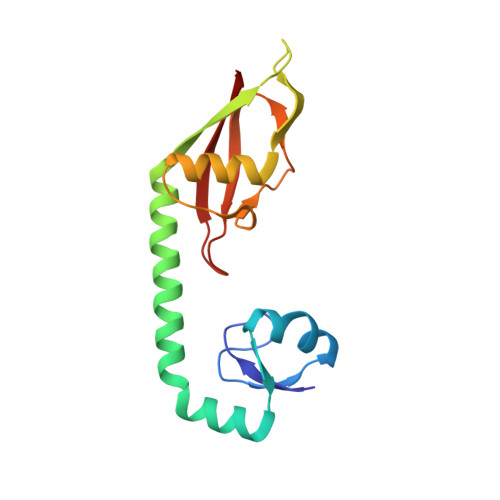 |
UniProt | |||||
Find proteins for P0A7R1 (Escherichia coli (strain K12)) Explore P0A7R1 Go to UniProtKB: P0A7R1 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P0A7R1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 16S rRNA | A [auth 2] | 1,542 | Escherichia coli | 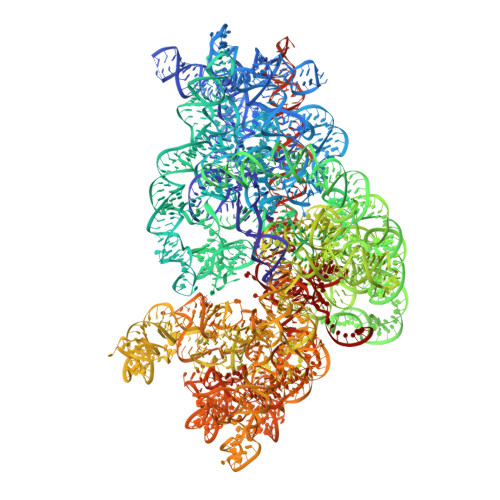 | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 22 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| mRNA | V [auth 4] | 49 | Escherichia coli |  | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 23 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| tRNA(fMet) | W [auth 5] | 76 | Escherichia coli | 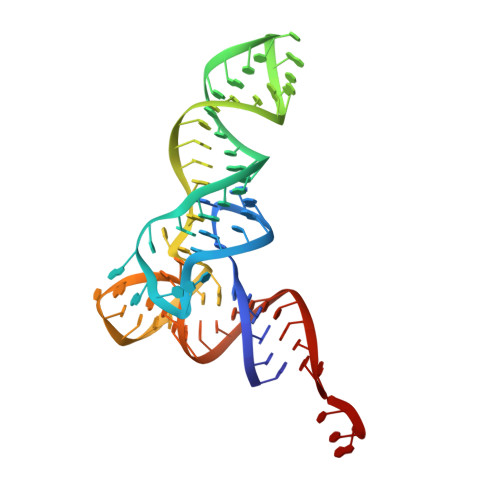 | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 24 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 23S rRNA | X [auth 1] | 2,904 | Escherichia coli |  | |
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 25 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| 5S rRNA | Y [auth 3] | 120 | Escherichia coli |  | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 6 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| SPM Query on SPM | SO [auth 1] | SPERMINE C10 H26 N4 PFNFFQXMRSDOHW-UHFFFAOYSA-N |  | ||
| SPD Query on SPD | IO [auth 1] JO [auth 1] KO [auth 1] KT [auth 1] LO [auth 1] | SPERMIDINE C7 H19 N3 ATHGHQPFGPMSJY-UHFFFAOYSA-N |  | ||
| PUT Query on PUT | AT [auth 1] BT [auth 1] CT [auth 1] DT [auth 1] EG [auth 2] | 1,4-DIAMINOBUTANE C4 H12 N2 KIDHWZJUCRJVML-UHFFFAOYSA-N |  | ||
| ZN Query on ZN | VT [auth f], WT [auth a] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| K Query on K | AF [auth 2] AP [auth 1] AQ [auth 1] AR [auth 1] AS [auth 1] | POTASSIUM ION K NPYPAHLBTDXSSS-UHFFFAOYSA-N |  | ||
| MG Query on MG | AC [auth 2] AD [auth 2] AE [auth 2] AG [auth 2] AH [auth 1] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Modified Residues 4 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| D2T Query on D2T | L [auth q] | L-PEPTIDE LINKING | C5 H9 N O4 S |  | ASP |
| MEQ Query on MEQ | AA [auth C] | L-PEPTIDE LINKING | C6 H12 N2 O3 |  | GLN |
| 4D4 Query on 4D4 | HA [auth M] | L-PEPTIDE LINKING | C6 H14 N4 O3 |  | ARG |
| MS6 Query on MS6 | HA [auth M] | L-PEPTIDE LINKING | C5 H11 N O S2 |  | MET |
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cryoSPARC | 4.2 |
| MODEL REFINEMENT | PHENIX | 1.20 |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Swedish Research Council | Sweden | 2016-06264 |
| Swedish Research Council | Sweden | 2018-05946 |
| Swedish Research Council | Sweden | 2018-05498 |
| Knut and Alice Wallenberg Foundation | Sweden | KAW 2017.0055 |
| Carl Trygger Foundation | Sweden | CTS 18:338 |
| Carl Trygger Foundation | Sweden | CTS 19:806 |
| Wenner-Gren Foundation | United States | UPD2017:0238 |
| Swedish Research Council | Sweden | 2016-06264 |
| Swedish Research Council | Sweden | 2017-03827 |