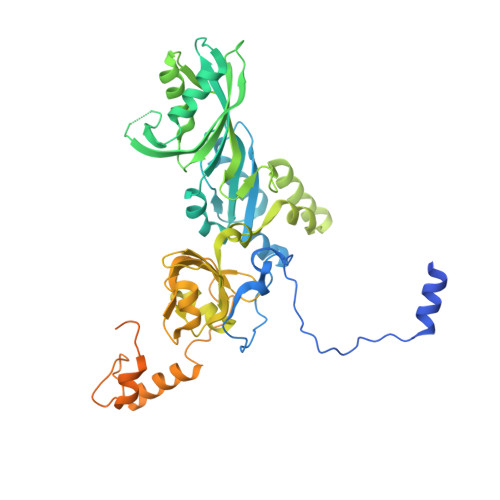
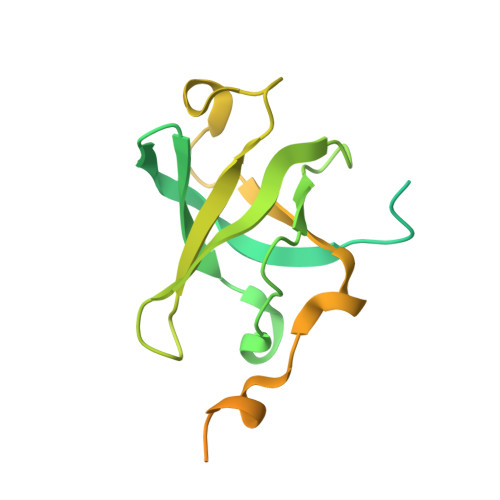
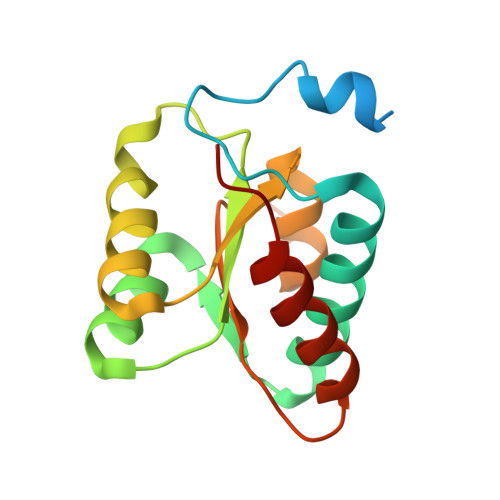
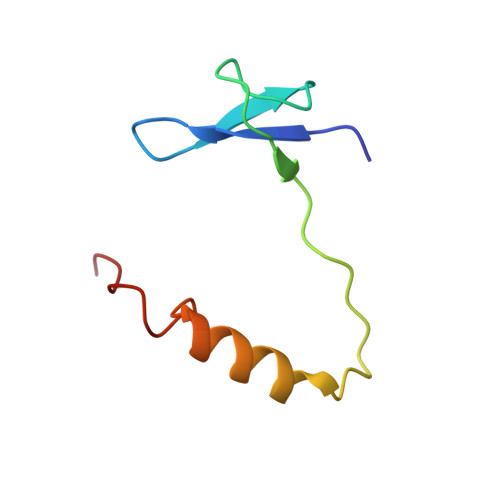
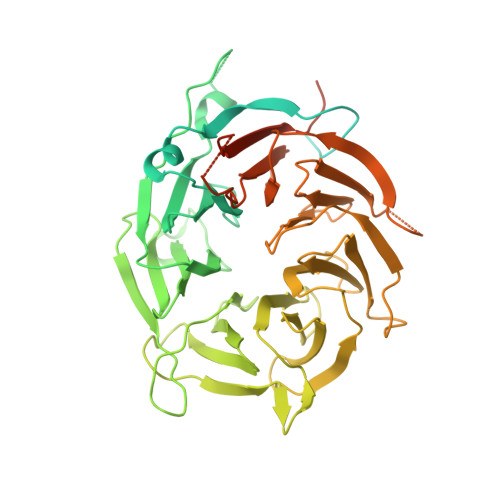
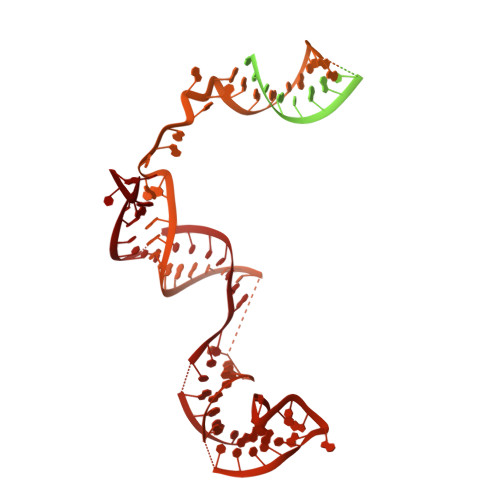
2.7 angstrom cryo-EM structure of human telomerase H/ACA ribonucleoprotein.
Ghanim, G.E., Sekne, Z., Balch, S., van Roon, A.M., Nguyen, T.H.D.(2024) Nat Commun 15: 746-746
- PubMed: 38272871
- DOI: https://doi.org/10.1038/s41467-024-45002-x
- Primary Citation of Related Structures:
8OUE, 8OUF - PubMed Abstract:
Telomerase is a ribonucleoprotein (RNP) enzyme that extends telomeric repeats at eukaryotic chromosome ends to counterbalance telomere loss caused by incomplete genome replication. Human telomerase is comprised of two distinct functional lobes tethered by telomerase RNA (hTR): a catalytic core, responsible for DNA extension; and a Hinge and ACA (H/ACA) box RNP, responsible for telomerase biogenesis. H/ACA RNPs also have a general role in pseudouridylation of spliceosomal and ribosomal RNAs, which is critical for the biogenesis of the spliceosome and ribosome. Much of our structural understanding of eukaryotic H/ACA RNPs comes from structures of the human telomerase H/ACA RNP. Here we report a 2.7 Å cryo-electron microscopy structure of the telomerase H/ACA RNP. The significant improvement in resolution over previous 3.3 Å to 8.2 Å structures allows us to uncover new molecular interactions within the H/ACA RNP. Many disease mutations are mapped to these interaction sites. The structure also reveals unprecedented insights into a region critical for pseudouridylation in canonical H/ACA RNPs. Together, our work advances understanding of telomerase-related disease mutations and the mechanism of pseudouridylation by eukaryotic H/ACA RNPs.
- MRC Laboratory of Molecular Biology, Cambridge, CB2 0QH, UK.
Organizational Affiliation: