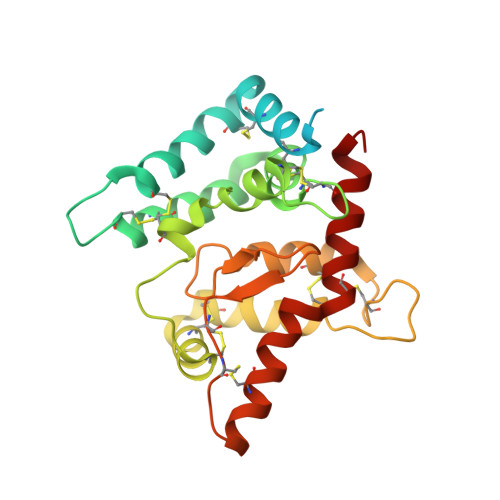
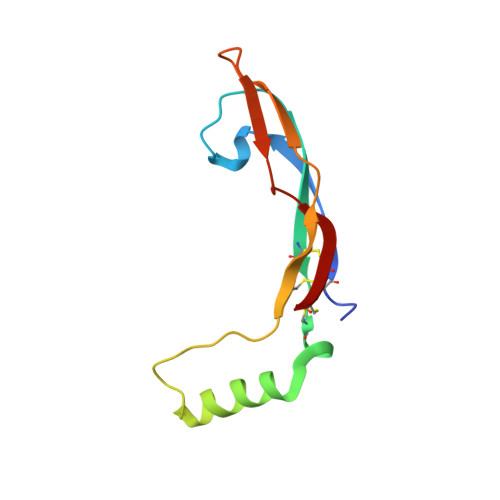
Architecture and regulation of a GDNF-GFR alpha 1 synaptic adhesion assembly.
Houghton, F.M., Adams, S.E., Rios, A.S., Masino, L., Purkiss, A.G., Briggs, D.C., Ledda, F., McDonald, N.Q.(2023) Nat Commun 14: 7551-7551
- PubMed: 37985758
- DOI: https://doi.org/10.1038/s41467-023-43148-8
- Primary Citation of Related Structures:
8OS6 - PubMed Abstract:
Glial-cell line derived neurotrophic factor (GDNF) bound to its co-receptor GFRα1 stimulates the RET receptor tyrosine kinase, promoting neuronal survival and neuroprotection. The GDNF-GFRα1 complex also supports synaptic cell adhesion independently of RET. Here, we describe the structure of a decameric GDNF-GFRα1 assembly determined by crystallography and electron microscopy, revealing two GFRα1 pentamers bridged by five GDNF dimers. We reconsitituted the assembly between adhering liposomes and used cryo-electron tomography to visualize how the complex fulfils its membrane adhesion function. The GFRα1:GFRα1 pentameric interface was further validated both in vitro by native PAGE and in cellulo by cell-clustering and dendritic spine assays. Finally, we provide biochemical and cell-based evidence that RET and heparan sulfate cooperate to prevent assembly of the adhesion complex by competing for the adhesion interface. Our results provide a mechanistic framework to understand GDNF-driven cell adhesion, its relationship to trophic signalling, and the central role played by GFRα1.
- Signalling and Structural Biology laboratory, The Francis Crick Institute, 1 Midland Road, London, NW1 1AT, UK.
Organizational Affiliation: