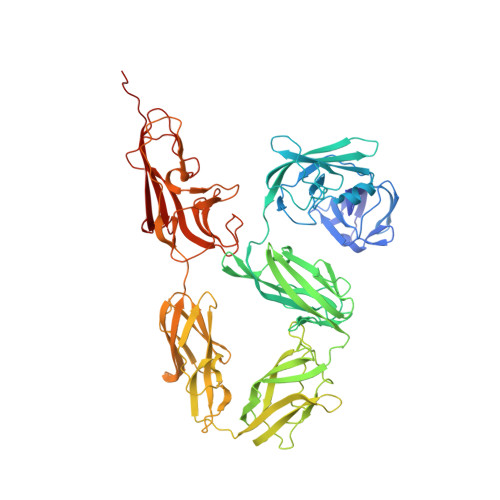
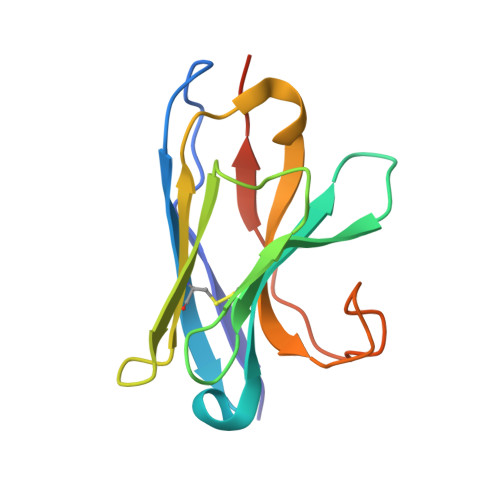
Structure and function of the EA1 surface layer of Bacillus anthracis.
Sogues, A., Fioravanti, A., Jonckheere, W., Pardon, E., Steyaert, J., Remaut, H.(2023) Nat Commun 14: 7051-7051
- PubMed: 37923757
- DOI: https://doi.org/10.1038/s41467-023-42826-x
- Primary Citation of Related Structures:
8OPR - PubMed Abstract:
The Gram-positive spore-forming bacterium Bacillus anthracis is the causative agent of anthrax, a deadly disease mostly affecting wildlife and livestock, as well as representing a bioterrorism threat. Its cell surface is covered by the mutually exclusive S-layers Sap and EA1, found in early and late growth phases, respectively. Here we report the nanobody-based structural characterization of EA1 and its native lattice contacts. The EA1 assembly domain consists of 6 immunoglobulin-like domains, where three calcium-binding sites structure interdomain contacts that allow monomers to adopt their assembly-competent conformation. Nanobody-induced depolymerization of EA1 S-layers results in surface defects, membrane blebbing and cell lysis under hypotonic conditions, indicating that S-layers provide additional mechanical stability to the cell wall. Taken together, we report a complete model of the EA1 S-layer and present a set of nanobodies that may have therapeutic potential against Bacillus anthracis.
- Structural and Molecular Microbiology, VIB-VUB Center for Structural Biology, VIB, Pleinlaan 2, 1050, Brussels, Belgium. Adria.Sogues.Castrejon@vub.be.
Organizational Affiliation: