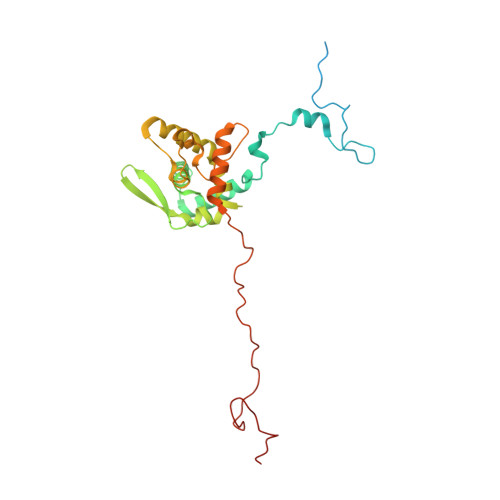
From structural polymorphism to structural metamorphosis of the coat protein of flexuous filamentous potato virus Y.
Kavcic, L., Kezar, A., Koritnik, N., Znidaric, M.T., Klobucar, T., Vicic, Z., Merzel, F., Holden, E., Benesch, J.L.P., Podobnik, M.(2024) Commun Chem 7: 14-14
- PubMed: 38233506
- DOI: https://doi.org/10.1038/s42004-024-01100-x
- Primary Citation of Related Structures:
8OPA, 8OPB, 8OPC, 8OPD, 8OPE, 8OPF, 8OPG, 8OPH, 8OPJ, 8OPK, 8OPL - PubMed Abstract:
The structural diversity and tunability of the capsid proteins (CPs) of various icosahedral and rod-shaped viruses have been well studied and exploited in the development of smart hybrid nanoparticles. However, the potential of CPs of the wide-spread flexuous filamentous plant viruses remains to be explored. Here, we show that we can control the shape, size, RNA encapsidation ability, symmetry, stability and surface functionalization of nanoparticles through structure-based design of CP from potato virus Y (PVY). We provide high-resolution insight into CP-based self-assemblies, ranging from large polymorphic or monomorphic filaments to smaller annular, cubic or spherical particles. Furthermore, we show that we can prevent CP self-assembly in bacteria by fusion with a cleavable protein, enabling controlled nanoparticle formation in vitro. Understanding the remarkable structural diversity of PVY CP not only provides possibilities for the production of biodegradable nanoparticles, but may also advance future studies of CP's polymorphism in a biological context.
- Department of Molecular Biology and Nanobiotechnology, National Institute of Chemistry, Ljubljana, Slovenia.
Organizational Affiliation: