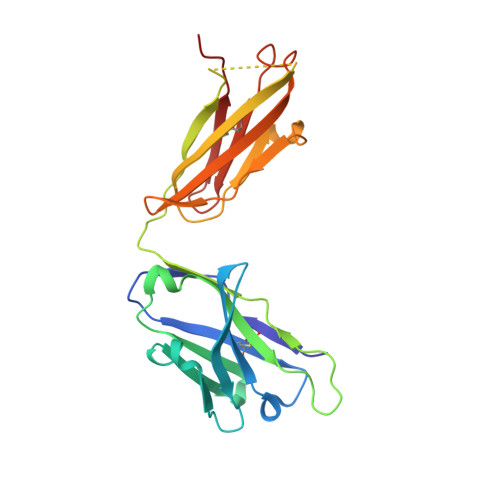
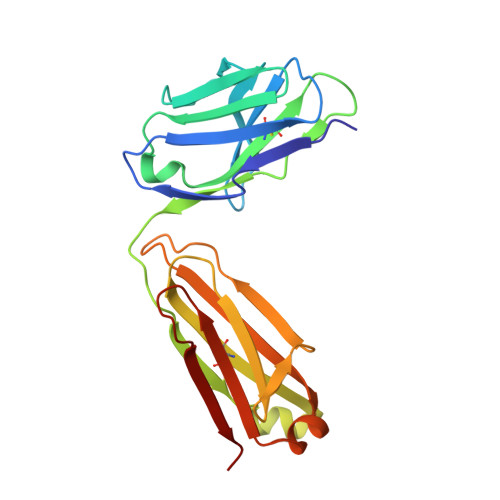
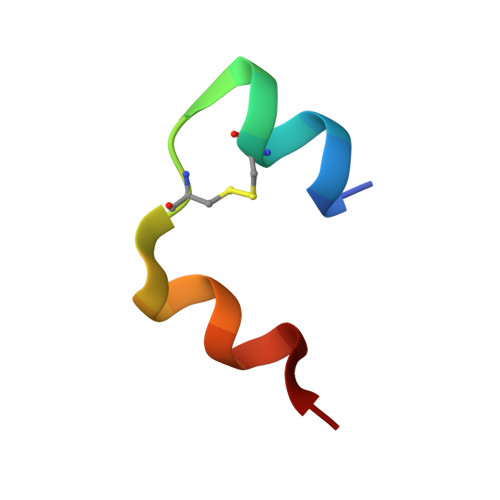
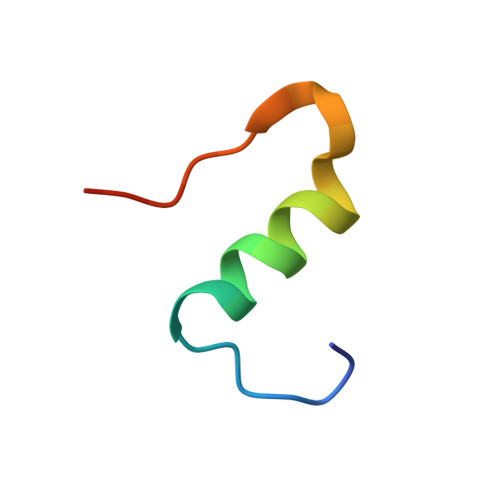
IgE clonality and aggregation of insulin affect IgE-mediated activation of sensitized basophils.
Christoffersen, S., Baumann, K., Millner, A.H., Toft-Hansen, H., Johansson, E., Thorlaksen, C., Skov, P.S.(2025) J Allergy Clin Immunol Glob 4: 100502-100502
- PubMed: 40583959
- DOI: https://doi.org/10.1016/j.jacig.2025.100502
- Primary Citation of Related Structures:
8ONI, 8ONK - PubMed Abstract:
Skin prick testing and specific IgE determination do not discriminate between sensitization and clinically relevant insulin allergic reactions, whereas the basophil histamine release assay has proven useful in identifying clinically relevant food and drug allergic reactions. We investigated IgE-mediated allergic reactions to insulin by studying the effect of insulin dimerization, superstructures (aggregates), and IgE clonality on activation of sensitized basophils. Three humanized IgE molecules directed against distinct insulin epitopes were developed, and basophils, alone or in combination, were sensitized with these antibodies. Insulin in monomeric or dimeric form and superstructures in solution or coupled to a Sepharose matrix were used to stimulate histamine release. Different insulin structures were tested on basophils sensitized with sera from 3 diabetic patients with allergic reactions and insulin-specific IgE during insulin treatment. Monoclonal humanized IgE molecules triggered basophil histamine release when insulin was dimerized, presented in a superstructure, or coupled to a Sepharose matrix-but not when insulin was monomeric. Monomeric insulin only induced histamine release when basophils were sensitized with a specific pair of IgE molecules. In basophils sensitized with sera from insulin-allergic patients containing insulin-specific IgE, only insulin in dimer formation, presented in a superstructure or coupled to a Sepharose matrix, could induce robust histamine release. Free monomeric insulin could not. This study illustrates why diagnostic methods for insulin allergy are difficult to correlate to clinical symptoms. Insulin aggregation and IgE clonality may be important factors in developing clinical symptoms of insulin allergy and should be addressed to improve the diagnostic outcome of insulin allergy testing.
- RefLab ApS, Hvidovre, Denmark.
Organizational Affiliation: