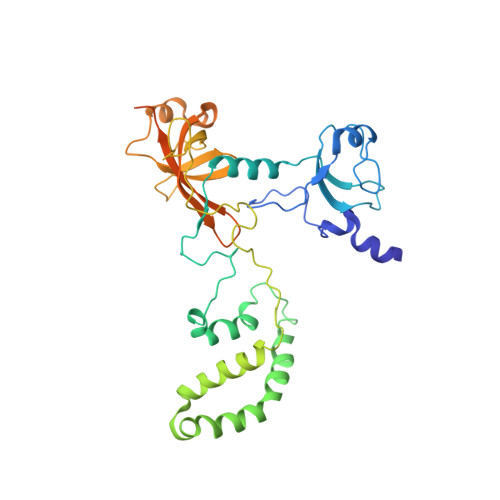
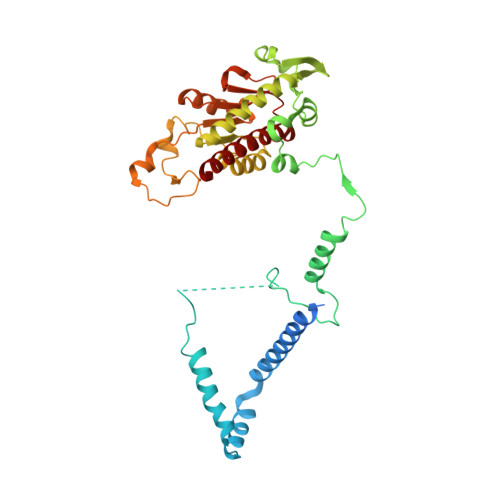
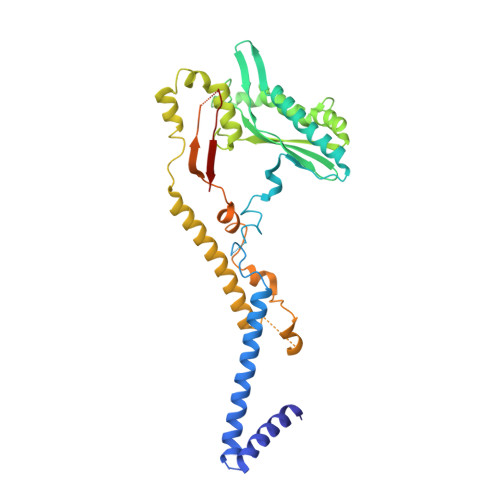
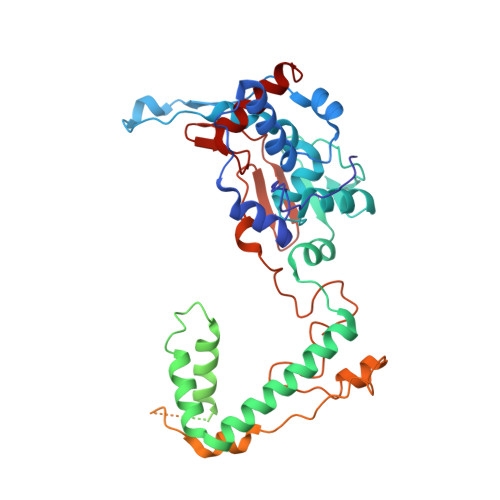
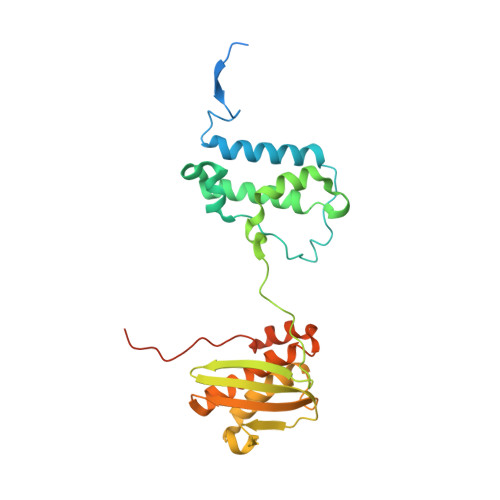
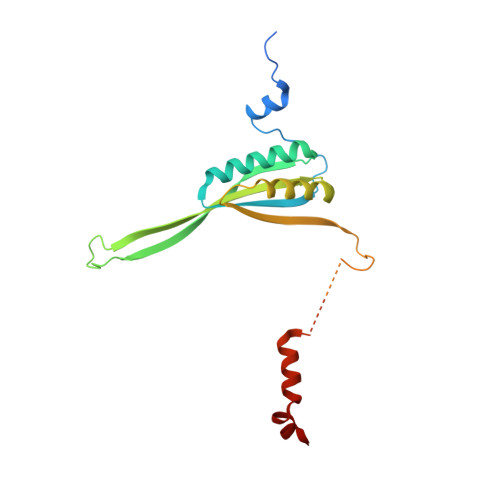
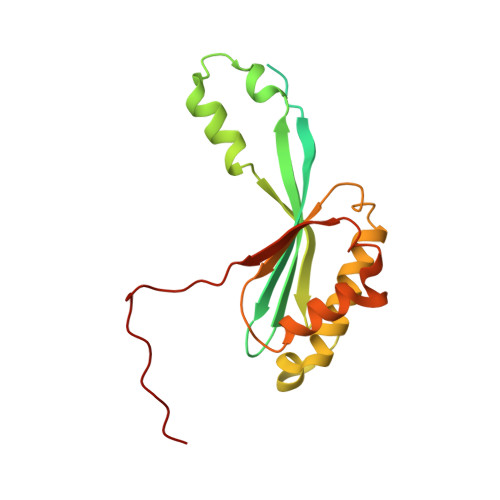
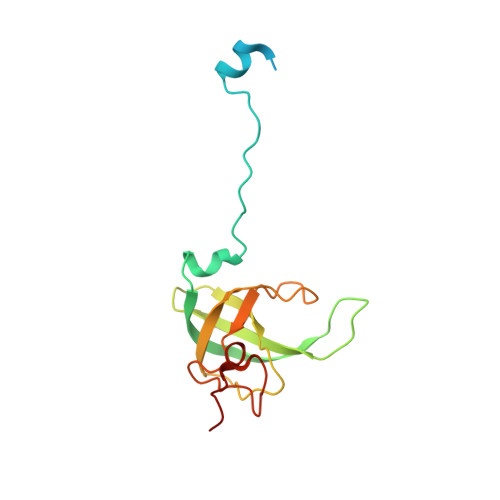
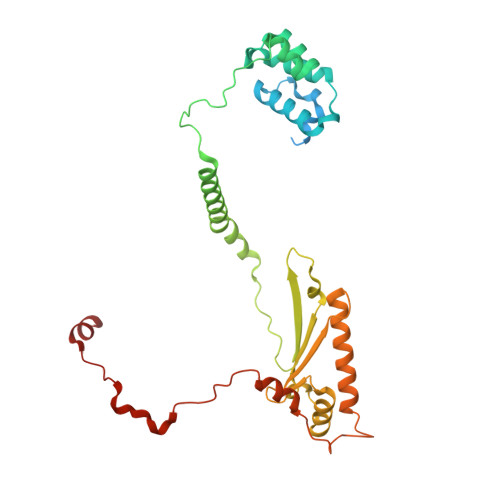
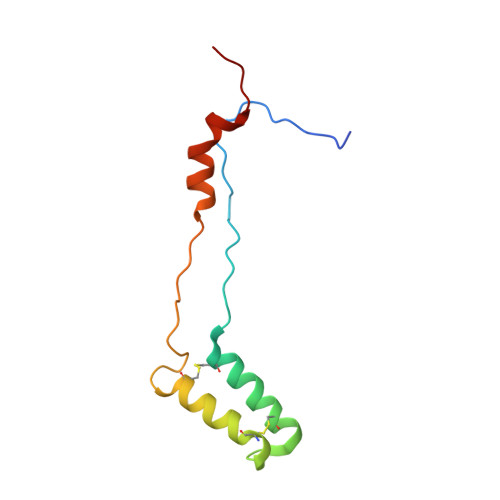
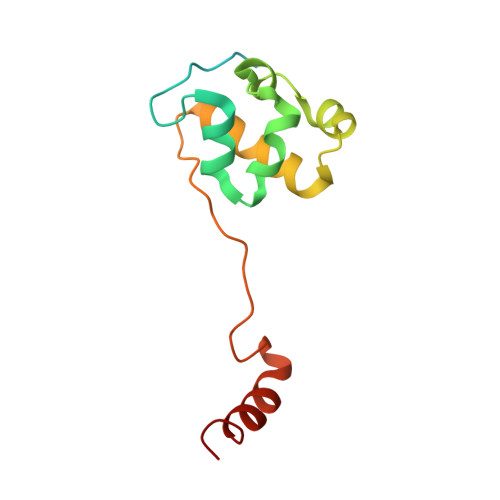
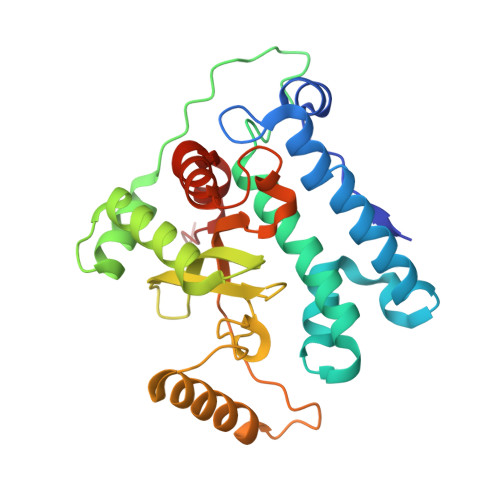
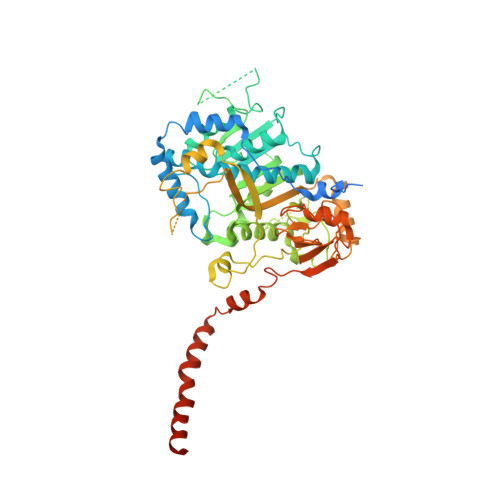
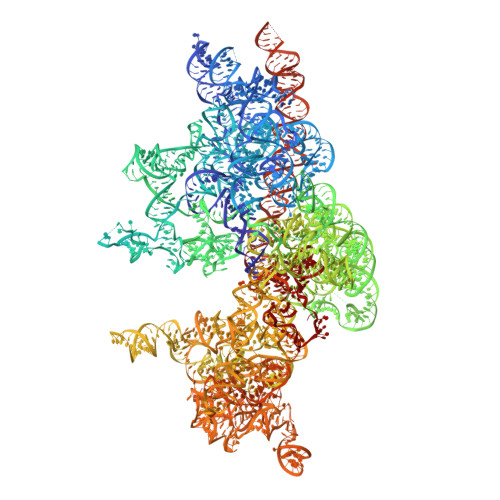
METTL17 is an Fe-S cluster checkpoint for mitochondrial translation.
Ast, T., Itoh, Y., Sadre, S., McCoy, J.G., Namkoong, G., Wengrod, J.C., Chicherin, I., Joshi, P.R., Kamenski, P., Suess, D.L.M., Amunts, A., Mootha, V.K.(2024) Mol Cell 84: 359
- PubMed: 38199006
- DOI: https://doi.org/10.1016/j.molcel.2023.12.016
- Primary Citation of Related Structures:
8OM2, 8OM3, 8OM4 - PubMed Abstract:
Friedreich's ataxia (FA) is a debilitating, multisystemic disease caused by the depletion of frataxin (FXN), a mitochondrial iron-sulfur (Fe-S) cluster biogenesis factor. To understand the cellular pathogenesis of FA, we performed quantitative proteomics in FXN-deficient human cells. Nearly every annotated Fe-S cluster-containing protein was depleted, indicating that as a rule, cluster binding confers stability to Fe-S proteins. We also observed depletion of a small mitoribosomal assembly factor METTL17 and evidence of impaired mitochondrial translation. Using comparative sequence analysis, mutagenesis, biochemistry, and cryoelectron microscopy, we show that METTL17 binds to the mitoribosomal small subunit during late assembly and harbors a previously unrecognized [Fe 4 S 4 ] 2+ cluster required for its stability. METTL17 overexpression rescued the mitochondrial translation and bioenergetic defects, but not the cellular growth, of FXN-depleted cells. These findings suggest that METTL17 acts as an Fe-S cluster checkpoint, promoting translation of Fe-S cluster-rich oxidative phosphorylation (OXPHOS) proteins only when Fe-S cofactors are replete.
- Broad Institute, Cambridge, MA 02142, USA; Howard Hughes Medical Institute, Massachusetts General Hospital, Boston, MA 02114, USA; Department of Molecular Biology, Massachusetts General Hospital, Boston, MA 02114, USA; Department of Systems Biology, Harvard Medical School, Boston, MA 02115, USA.
Organizational Affiliation: