Interrogation of an Enzyme Library Reveals the Catalytic Plasticity of Naturally Evolved [4+2] Cyclases.
Zorn, K., Back, C.R., Barringer, R., Chadimova, V., Manzo-Ruiz, M., Mbatha, S.Z., Mobarec, J.C., Williams, S.E., van der Kamp, M.W., Race, P.R., Willis, C.L., Hayes, M.A.(2023) Chembiochem 24: e202300382-e202300382
- PubMed: 37305956
- DOI: https://doi.org/10.1002/cbic.202300382
- Primary Citation of Related Structures:
8OF7 - PubMed Abstract:
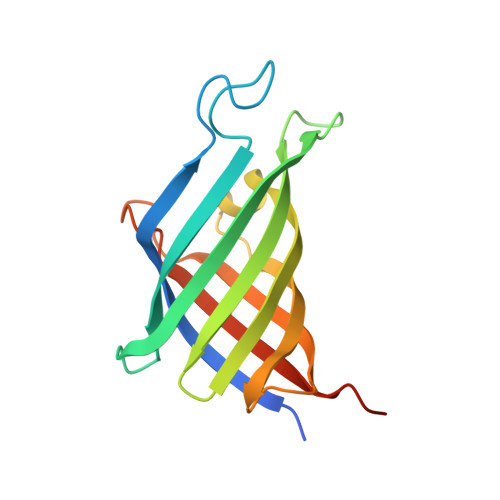
Stereoselective carbon-carbon bond forming reactions are quintessential transformations in organic synthesis. One example is the Diels-Alder reaction, a [4+2] cycloaddition between a conjugated diene and a dienophile to form cyclohexenes. The development of biocatalysts for this reaction is paramount for unlocking sustainable routes to a plethora of important molecules. To obtain a comprehensive understanding of naturally evolved [4+2] cyclases, and to identify hitherto uncharacterised biocatalysts for this reaction, we constructed a library comprising forty-five enzymes with reported or predicted [4+2] cycloaddition activity. Thirty-one library members were successfully produced in recombinant form. In vitro assays employing a synthetic substrate incorporating a diene and a dienophile revealed broad-ranging cycloaddition activity amongst these polypeptides. The hypothetical protein Cyc15 was found to catalyse an intramolecular cycloaddition to generate a novel spirotetronate. The crystal structure of this enzyme, along with docking studies, establishes the basis for stereoselectivity in Cyc15, as compared to other spirotetronate cyclases.
- Compound Synthesis and Management, Discovery Sciences, Biopharmaceuticals R&D, AstraZeneca, Pepparedsleden 1, 431 83, Mölndal, Sweden.
Organizational Affiliation: