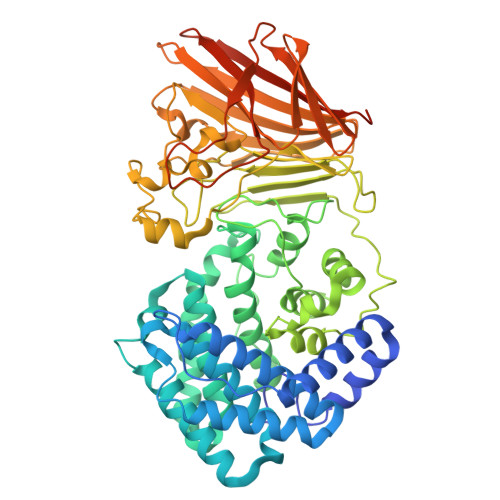
Crystal structure and catalytic mechanism of PL35 family glycosaminoglycan lyases with an ultrabroad substrate spectrum.
Wei, L., Cao, H.Y., Zou, R., Du, M., Zhang, Q., Lu, D., Xu, X., Xu, Y., Wang, W., Chen, X.L., Zhang, Y.Z., Li, F.(2025) Elife 13
- PubMed: 40387079
- DOI: https://doi.org/10.7554/eLife.102422
- Primary Citation of Related Structures:
8KHV - PubMed Abstract:
Recently, a new class of glycosaminoglycan (GAG) lyases (GAGases) belonging to PL35 family has been discovered with an ultrabroad substrate spectrum that can degrade three types of uronic acid-containing GAGs (hyaluronic acid, chondroitin sulfate and heparan sulfate) or even alginate. In this study, the structures of GAGase II from Spirosoma fluviale and GAGase VII from Bacteroides intestinalis DSM 17393 were determined at 1.9 and 2.4 Å resolution, respectively, and their catalytic mechanism was investigated by the site-directed mutant of their crucial residues and molecular docking assay. Structural analysis showed that GAGase II and GAGase VII consist of an N-terminal (α/α) 6 toroid multidomain and a C-terminal two-layered β-sheet domain with Mn 2+ . Notably, although GAGases share similar folds and catalytic mechanisms with some GAG lyases and alginate lyases, they exhibit higher structural similarity with alginate lyases than GAG lyases, which may present a crucial structural evidence for the speculation that GAG lyases with (α/α) n toroid and antiparallel β-sheet structures arrived by a divergent evolution from alginate lyases with the same folds. Overall, this study not only solved the structure of PL35 GAG lyases for the first time and investigated their catalytic mechanism, especially the reason why GAGase III can additionally degrade alginate, but also provided a key clue in the divergent evolution of GAG lyases that originated from alginate lyases.
- National Glycoengineering Research Center and Shandong Key Laboratory of Carbohydrate Chemistry and Glycobiology, State Key Laboratory of Microbial Technology, Shandong University, Qingdao, China.
Organizational Affiliation: