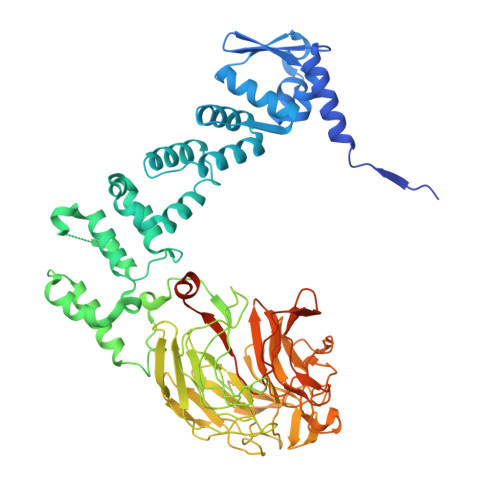
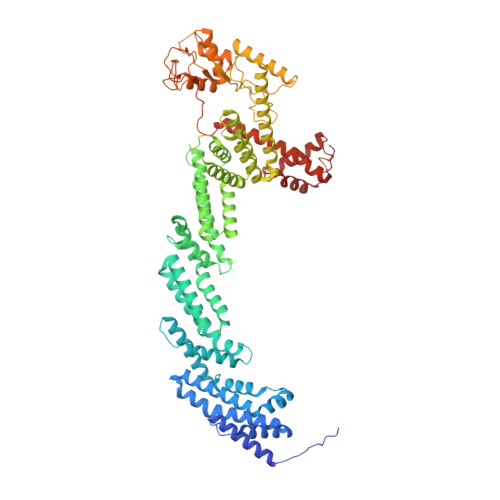
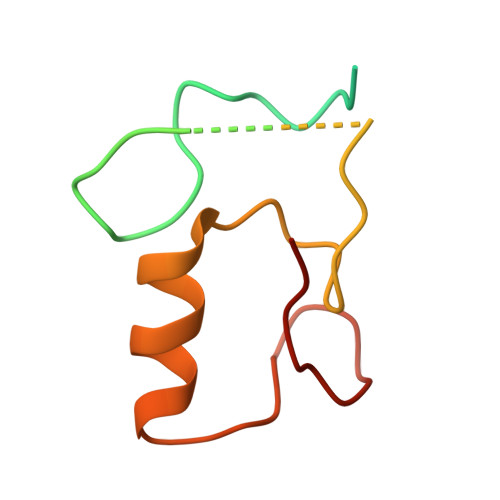
Cryo-EM structure of the KLHL22 E3 ligase bound to an oligomeric metabolic enzyme.
Teng, F., Wang, Y., Liu, M., Tian, S., Stjepanovic, G., Su, M.Y.(2023) Structure 31: 1431-1440.e5
- PubMed: 37788672
- DOI: https://doi.org/10.1016/j.str.2023.09.002
- Primary Citation of Related Structures:
8KGY, 8KHP, 8W4J - PubMed Abstract:
CULLIN-RING ligases constitute the largest group of E3 ubiquitin ligases. While some CULLIN family members recruit adapters before engaging further with different substrate receptors, homo-dimeric BTB-Kelch family proteins combine adapter and substrate receptor into a single polypeptide for the CULLIN3 family. However, the entire structural assembly and molecular details have not been elucidated to date. Here, we present a cryo-EM structure of the CULLIN3 RBX1 in complex with Kelch-like protein 22 (KLHL22) and a mitochondrial glutamate dehydrogenase complex I (GDH1) at 3.06 Å resolution. The structure adopts a W-shaped architecture formed by E3 ligase dimers. Three CULLIN3 KLHL22-RBX1 dimers were found to be dynamically associated with a single GDH1 hexamer. CULLIN3 KLHL22-RBX1 ligase mediated the polyubiquitination of GDH1 in vitro. Together, these results enabled the establishment of a structural model for understanding the complete assembly of BTB-Kelch proteins with CULLIN3 and how together they recognize oligomeric substrates and target them for ubiquitination.
- Department of Biochemistry, School of Medicine, Southern University of Science and Technology, Shenzhen 518055, China; Kobilka Institute of Innovative Drug Discovery, School of Medicine, The Chinese University of Hong Kong, Shenzhen, Shenzhen 518172, China.
Organizational Affiliation: