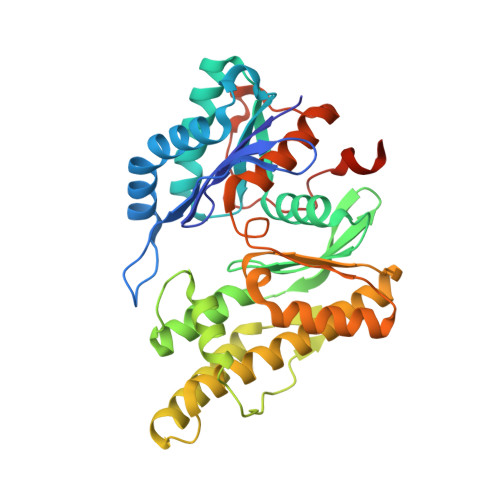
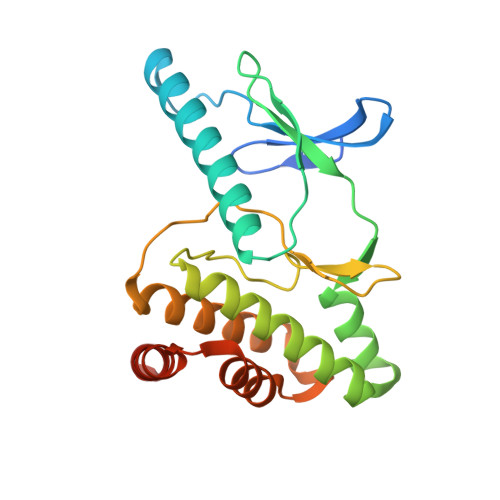
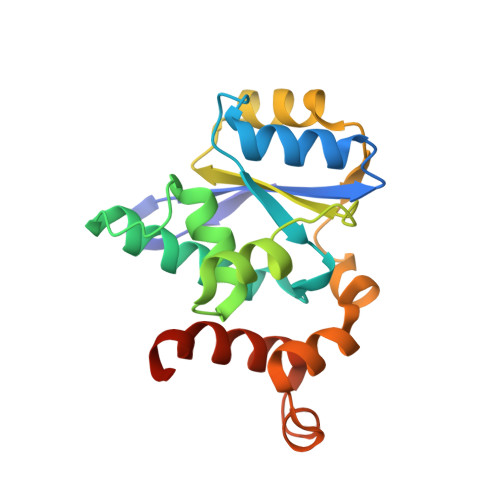
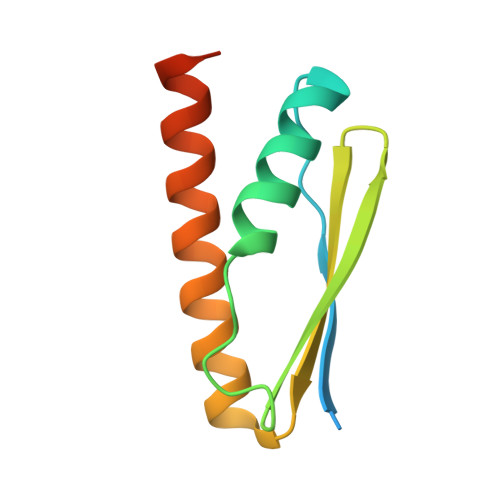
Molecular basis of A. thaliana KEOPS complex in biosynthesizing tRNA t6A.
Zheng, X., Su, C., Duan, L., Jin, M., Sun, Y., Zhu, L., Zhang, W.(2024) Nucleic Acids Res 52: 4523-4540
- PubMed: 38477398
- DOI: https://doi.org/10.1093/nar/gkae179
- Primary Citation of Related Structures:
8K20 - PubMed Abstract:
In archaea and eukaryotes, the evolutionarily conserved KEOPS is composed of four core subunits-Kae1, Bud32, Cgi121 and Pcc1, and a fifth Gon7/Pcc2 that is found in fungi and metazoa. KEOPS cooperates with Sua5/YRDC to catalyze the biosynthesis of tRNA N6-threonylcarbamoyladenosine (t6A), an essential modification needed for fitness of cellular organisms. Biochemical and structural characterizations of KEOPSs from archaea, yeast and humans have determined a t6A-catalytic role for Kae1 and auxiliary roles for other subunits. However, the precise molecular workings of KEOPSs still remain poorly understood. Here, we investigated the biochemical functions of A. thaliana KEOPS and determined a cryo-EM structure of A. thaliana KEOPS dimer. We show that A. thaliana KEOPS is composed of KAE1, BUD32, CGI121 and PCC1, which adopts a conserved overall arrangement. PCC1 dimerization leads to a KEOPS dimer that is needed for an active t6A-catalytic KEOPS-tRNA assembly. BUD32 participates in direct binding of tRNA to KEOPS and modulates the t6A-catalytic activity of KEOPS via its C-terminal tail and ATP to ADP hydrolysis. CGI121 promotes the binding of tRNA to KEOPS and potentiates the t6A-catalytic activity of KEOPS. These data and findings provide insights into mechanistic understanding of KEOPS machineries.
- School of Life Sciences, Key Laboratory of Cell Activities and Stress Adaptation of the Ministry of Education, Lanzhou University, Lanzhou 730000, China.
Organizational Affiliation: