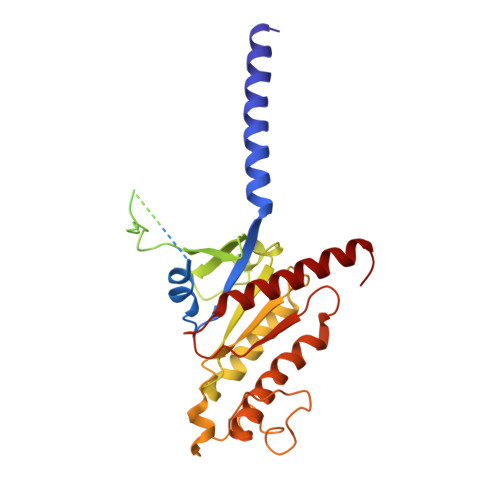
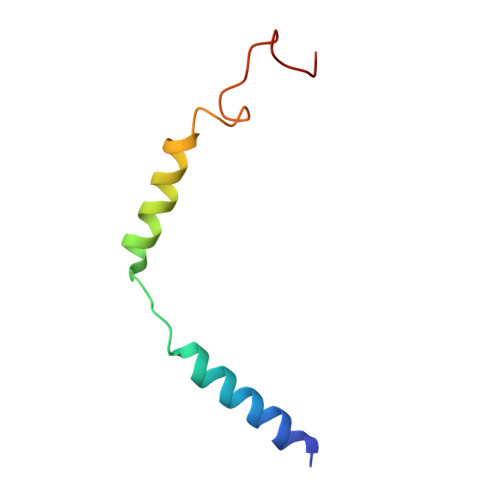
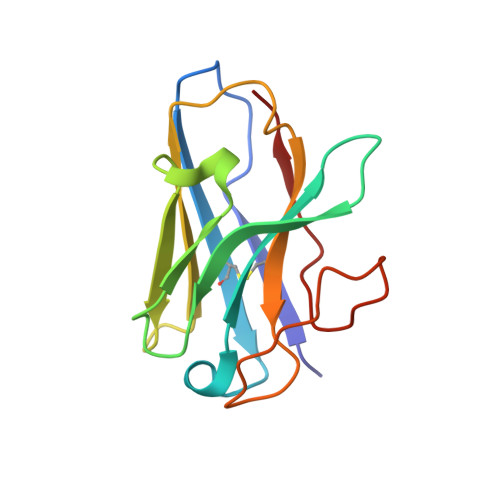
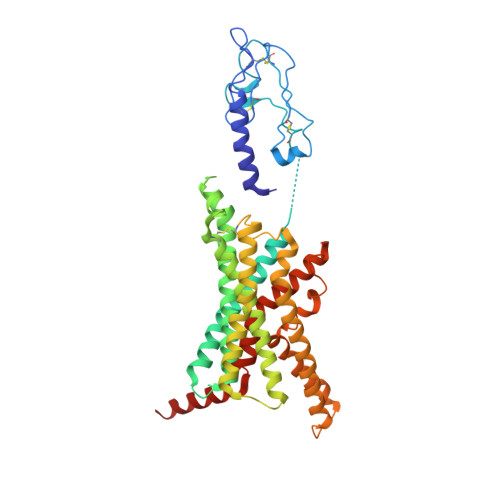
Structural analysis of the dual agonism at GLP-1R and GCGR.
Li, Y., Zhou, Q., Dai, A., Zhao, F., Chang, R., Ying, T., Wu, B., Yang, D., Wang, M.W., Cong, Z.(2023) Proc Natl Acad Sci U S A 120: e2303696120-e2303696120
- PubMed: 37549266
- DOI: https://doi.org/10.1073/pnas.2303696120
- Primary Citation of Related Structures:
8JIP, 8JIQ, 8JIR, 8JIS, 8JIT, 8JIU - PubMed Abstract:
Glucagon-like peptide-1 receptor (GLP-1R) and glucagon receptor (GCGR), two members of class B1 G protein-coupled receptors, play important roles in glucose homeostasis and energy metabolism. They share a high degree of sequence homology but have different functionalities. Unimolecular dual agonists of both receptors developed recently displayed better clinical efficacies than that of monotherapy. To study the underlying molecular mechanisms, we determined high-resolution cryo-electron microscopy structures of GLP-1R or GCGR in complex with heterotrimeric G s protein and three GLP-1R/GCGR dual agonists including peptide 15, MEDI0382 (cotadutide) and SAR425899 with variable activating profiles at GLP-1R versus GCGR. Compared with related structures reported previously and supported by our published pharmacological data, key residues responsible for ligand recognition and dual agonism were identified. Analyses of peptide conformational features revealed a difference in side chain orientations within the first three residues, indicating that distinct engagements in the deep binding pocket are required to achieve receptor selectivity. The middle region recognizes extracellular loop 1 (ECL1), ECL2, and the top of transmembrane helix 1 (TM1) resulting in specific conformational changes of both ligand and receptor, especially the dual agonists reshaped ECL1 conformation of GLP-1R relative to that of GCGR, suggesting an important role of ECL1 interaction in executing dual agonism. Structural investigation of lipid modification showed a better interaction between lipid moiety of MEDI0382 and TM1-TM2 cleft, in line with its increased potency at GCGR than SAR425899. Together, the results provide insightful information for the design and development of improved therapeutics targeting these two receptors simultaneously.
- Department of Medical Microbiology and Parasitology, Shanghai Institute of Infectious Disease and Biosecurity, School of Basic Medical Sciences, Fudan University, Shanghai 200032, China.
Organizational Affiliation: