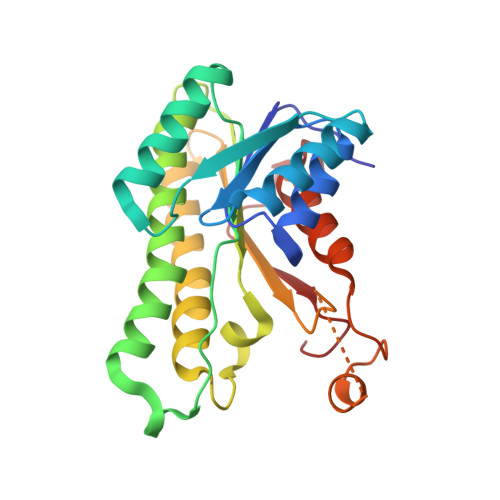
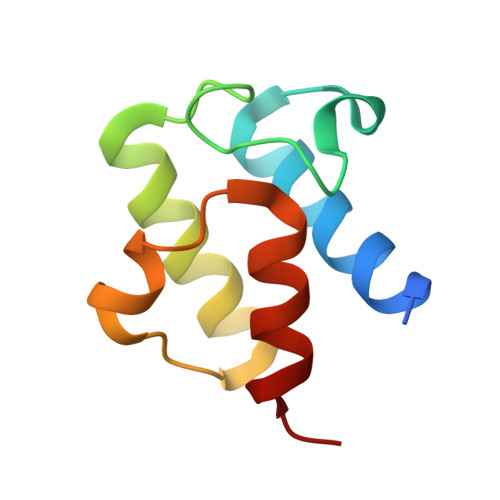
The Molecular Basis of Catalysis by SDR Family Members Ketoacyl-ACP Reductase FabG and Enoyl-ACP Reductase FabI in Type-II Fatty Acid Biosynthesis.
Zhou, J., Zhang, L., Wang, Y., Song, W., Huang, Y., Mu, Y., Schmitz, W., Zhang, S.Y., Lin, H., Chen, H.Z., Ye, F., Zhang, L.(2023) Angew Chem Int Ed Engl 62: e202313109-e202313109
- PubMed: 37779101
- DOI: https://doi.org/10.1002/anie.202313109
- Primary Citation of Related Structures:
8JF9, 8JFA, 8JFG, 8JFH, 8JFI, 8JFJ, 8JFM, 8JFN - PubMed Abstract:
The short-chain dehydrogenase/reductase (SDR) superfamily members acyl-ACP reductases FabG and FabI are indispensable core enzymatic modules and catalytic orientation controllers in type-II fatty acid biosynthesis. Herein, we report their distinct substrate allosteric recognition and enantioselective reduction mechanisms. FabG achieves allosteric regulation of ACP and NADPH through ACP binding across two adjacent FabG monomers, while FabI follows an irreversible compulsory order of substrate binding in that NADH binding must precede that of ACP on a discrete FabI monomer. Moreover, FabG and FabI utilize a backdoor residue Phe187 or a "rheostat" α8 helix for acyl chain length selection, and their corresponding triad residues Ser142 or Tyr145 recognize the keto- or enoyl-acyl substrates, respectively, facilitating initiation of nucleophilic attack by NAD(P)H. The other two triad residues (Tyr and Lys) mediate subsequent proton transfer and (R)-3-hydroxyacyl- or saturated acyl-ACP production.
- Department of Pharmacology and Chemical Biology, State Key Laboratory of Systems Medicine for Cancer, Shanghai Jiao Tong University School of Medicine, Shanghai, 200025, China.
Organizational Affiliation: