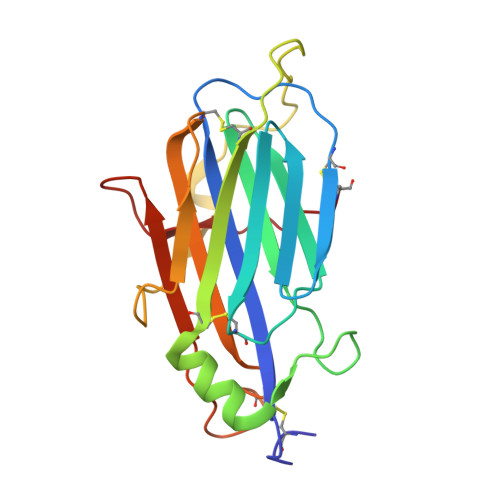
Expression, purification, and crystal structure of mpox virus A41 protein.
Jiang, H., Li, J., Jian, Y., Yang, T., Zhang, J., Li, J.(2024) Protein Expr Purif 219: 106480-106480
- PubMed: 38588871
- DOI: https://doi.org/10.1016/j.pep.2024.106480
- Primary Citation of Related Structures:
8JC6 - PubMed Abstract:
Mpox is a zoonotic disease that was once endemic in Africa countries caused by mpox virus. However, cases recently have been confirmed in many non-endemic countries outside of Africa. The rapidly increasing number of confirmed mpox cases poses a threat to the international community. In-depth studies of key viral factors are urgently needed, which will inform the design of multiple antiviral agents. Mpox virus A41L gene encodes a secreted protein, A41, that is nonessential for viral replication, but could affect the host response to infection via interacting with chemokines. Here, mpox virus A41 protein was expressed in Sf9 cells, and purified by affinity chromatography followed by gel filtration. Surface plasmon resonance spectroscopy showed that purified A41 binds a certain human chemokine CXCL8 with the equilibrium dissociation constant (K D ) being 1.22 × 10 -6 M. The crystal structure of mpox virus A41 protein was solved at 1.92 Å. Structural analysis and comparison revealed that mpox virus A41 protein adopts a characteristic β-sheet topology, showing minor differences with that of vaccinia virus. These preliminary structural and functional studies of A41 protein from mpox virus will help us better understand its role in chemokine subversion, and contributing to the knowledge to viral chemokine binding proteins.
- School of Basic Medical Sciences, Jiangxi Medical College, Nanchang University, Nanchang, 330031, China. Electronic address: haihaijiang2020@ncu.edu.cn.
Organizational Affiliation: