Molecular insights into atypical modes of beta-arrestin interaction with seven transmembrane receptors.
Maharana, J., Sano, F.K., Sarma, P., Yadav, M.K., Duan, L., Stepniewski, T.M., Chaturvedi, M., Ranjan, A., Singh, V., Saha, S., Mahajan, G., Chami, M., Shihoya, W., Selent, J., Chung, K.Y., Banerjee, R., Nureki, O., Shukla, A.K.(2024) Science 383: 101-108
- PubMed: 38175886
- DOI: https://doi.org/10.1126/science.adj3347
- Primary Citation of Related Structures:
8GO9, 8J8R, 8J8V, 8J8Z, 8J97, 8J9K, 8JA3, 8JAF - PubMed Abstract:
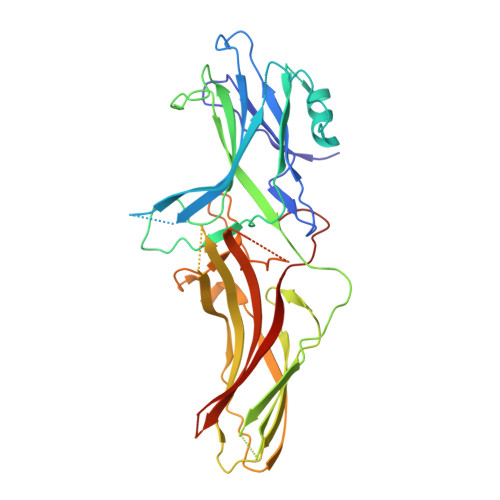
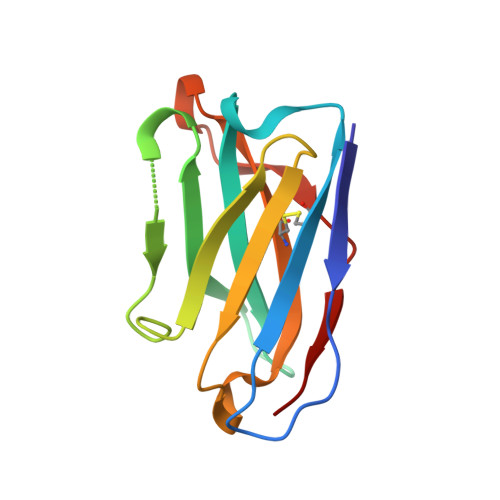
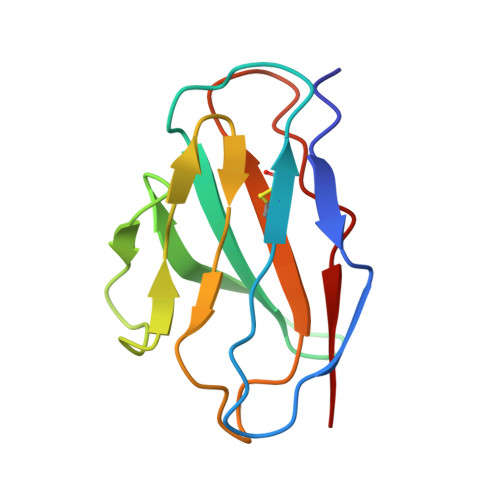
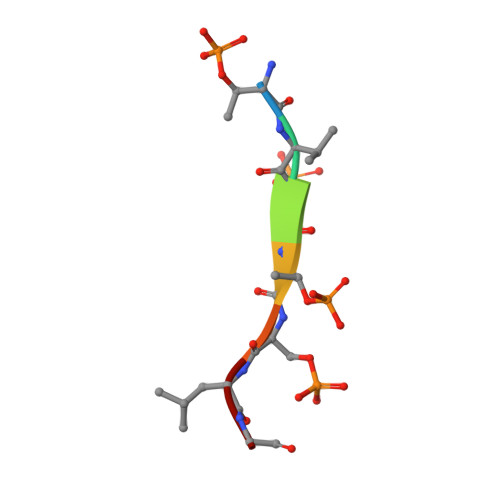
β-arrestins (βarrs) are multifunctional proteins involved in signaling and regulation of seven transmembrane receptors (7TMRs), and their interaction is driven primarily by agonist-induced receptor activation and phosphorylation. Here, we present seven cryo-electron microscopy structures of βarrs either in the basal state, activated by the muscarinic receptor subtype 2 (M2R) through its third intracellular loop, or activated by the βarr-biased decoy D6 receptor (D6R). Combined with biochemical, cellular, and biophysical experiments, these structural snapshots allow the visualization of atypical engagement of βarrs with 7TMRs and also reveal a structural transition in the carboxyl terminus of βarr2 from a β strand to an α helix upon activation by D6R. Our study provides previously unanticipated molecular insights into the structural and functional diversity encoded in 7TMR-βarr complexes with direct implications for exploring novel therapeutic avenues.
- Department of Biological Sciences, Indian Institute of Technology Kanpur, Kanpur, India.
Organizational Affiliation: