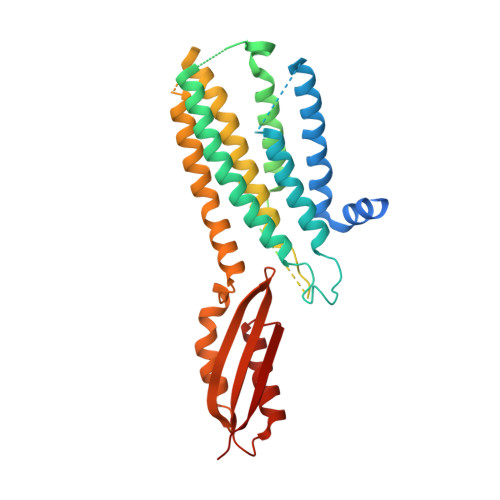
Cryo-EM structures of human zinc transporter ZnT7 reveal the mechanism of Zn 2+ uptake into the Golgi apparatus.
Bui, H.B., Watanabe, S., Nomura, N., Liu, K., Uemura, T., Inoue, M., Tsutsumi, A., Fujita, H., Kinoshita, K., Kato, Y., Iwata, S., Kikkawa, M., Inaba, K.(2023) Nat Commun 14: 4770-4770
- PubMed: 37553324
- DOI: https://doi.org/10.1038/s41467-023-40521-5
- Primary Citation of Related Structures:
8J7T, 8J7U, 8J7V, 8J7W, 8J7X, 8J7Y, 8J80 - PubMed Abstract:
Zinc ions (Zn 2+ ) are vital to most cells, with the intracellular concentrations of Zn 2+ being tightly regulated by multiple zinc transporters located at the plasma and organelle membranes. We herein present the 2.2-3.1 Å-resolution cryo-EM structures of a Golgi-localized human Zn 2+ /H + antiporter ZnT7 (hZnT7) in Zn 2+ -bound and unbound forms. Cryo-EM analyses show that hZnT7 exists as a dimer via tight interactions in both the cytosolic and transmembrane (TM) domains of two protomers, each of which contains a single Zn 2+ -binding site in its TM domain. hZnT7 undergoes a TM-helix rearrangement to create a negatively charged cytosolic cavity for Zn 2+ entry in the inward-facing conformation and widens the luminal cavity for Zn 2+ release in the outward-facing conformation. An exceptionally long cytosolic histidine-rich loop characteristic of hZnT7 binds two Zn 2+ ions, seemingly facilitating Zn 2+ recruitment to the TM metal transport pathway. These structures permit mechanisms of hZnT7-mediated Zn 2+ uptake into the Golgi to be proposed.
- Institute of Multidisciplinary Research for Advanced Materials, Tohoku University, Sendai, Miyagi, 980-8577, Japan.
Organizational Affiliation: