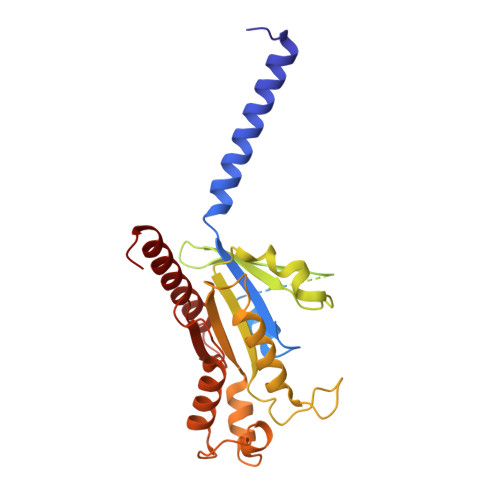
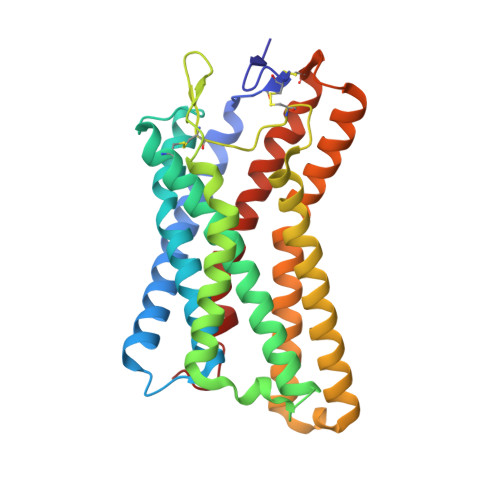
Orthosteric and allosteric modulation of human HCAR2 signaling complex.
Mao, C., Gao, M., Zang, S.K., Zhu, Y., Shen, D.D., Chen, L.N., Yang, L., Wang, Z., Zhang, H., Wang, W.W., Shen, Q., Lu, Y., Ma, X., Zhang, Y.(2023) Nat Commun 14: 7620-7620
- PubMed: 37993467
- DOI: https://doi.org/10.1038/s41467-023-43537-z
- Primary Citation of Related Structures:
8J6P, 8J6Q, 8J6R - PubMed Abstract:
Hydroxycarboxylic acids are crucial metabolic intermediates involved in various physiological and pathological processes, some of which are recognized by specific hydroxycarboxylic acid receptors (HCARs). HCAR2 is one such receptor, activated by endogenous β-hydroxybutyrate (3-HB) and butyrate, and is the target for Niacin. Interest in HCAR2 has been driven by its potential as a therapeutic target in cardiovascular and neuroinflammatory diseases. However, the limited understanding of how ligands bind to this receptor has hindered the development of alternative drugs able to avoid the common flushing side-effects associated with Niacin therapy. Here, we present three high-resolution structures of HCAR2-Gi1 complexes bound to four different ligands, one potent synthetic agonist (MK-6892) bound alone, and the two structures bound to the allosteric agonist compound 9n in conjunction with either the endogenous ligand 3-HB or niacin. These structures coupled with our functional and computational analyses further our understanding of ligand recognition, allosteric modulation, and activation of HCAR2 and pave the way for the development of high-efficiency drugs with reduced side-effects.
- Department of Biophysics and Department of Pathology of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, 310058, China. maochunyou@zju.edu.cn.
Organizational Affiliation: