Molecular recognition of niacin and lipid-lowering drugs by the human hydroxycarboxylic acid receptor 2.
Zhu, S., Yuan, Q., Li, X., He, X., Shen, S., Wang, D., Li, J., Cheng, X., Duan, X., Xu, H.E., Duan, J.(2023) Cell Rep 42: 113406-113406
- PubMed: 37952153
- DOI: https://doi.org/10.1016/j.celrep.2023.113406
- Primary Citation of Related Structures:
8J6I, 8J6J, 8J6L - PubMed Abstract:
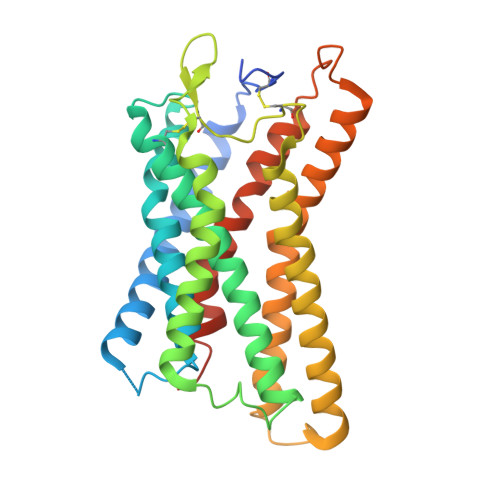
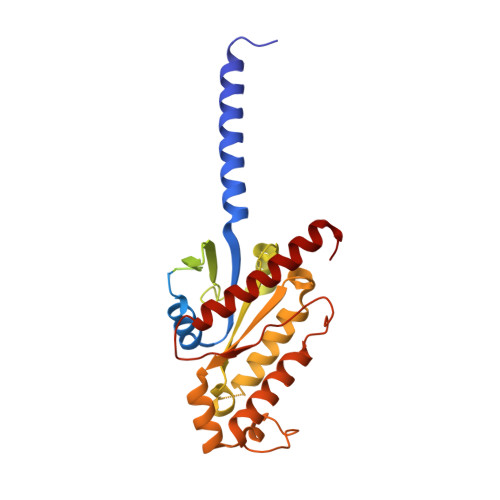
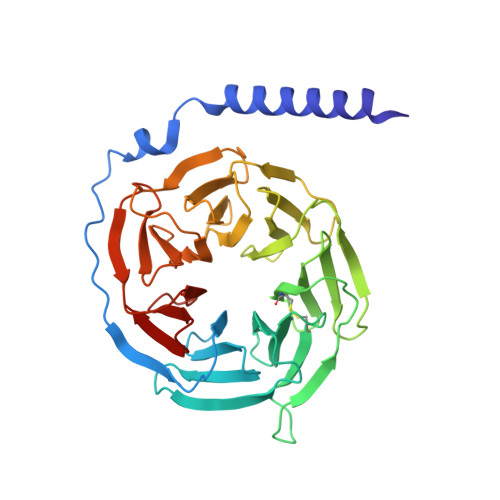
Niacin, an age-old lipid-lowering drug, acts through the hydroxycarboxylic acid receptor 2 (HCAR2), a G-protein-coupled receptor (GPCR). Yet, its use is hindered by side effects like skin flushing. To address this, specific HCAR2 agonists, like MK-6892 and GSK256073, with fewer adverse effects have been created. However, the activation mechanism of HCAR2 by niacin and these new agonists is not well understood. Here, we present three cryoelectron microscopy structures of Gi-coupled HCAR2 bound to niacin, MK-6892, and GSK256073. Our findings show that different ligands induce varying binding pockets in HCAR2, influenced by aromatic amino acid clusters (W91 ECL1 , H161 4.59 , W188 5.38 , H189 5.39 , and F193 5.43 ) from receptors ECL1, TM4, and TM5. Additionally, conserved residues R111 3.36 and Y284 7.43 , unique to the HCA receptor family, likely initiate activation signal propagation in HCAR2. This study provides insights into ligand recognition, receptor activation, and G protein coupling mediated by HCAR2, laying the groundwork for developing HCAR2-targeted drugs.
- School of Pharmacy, Faculty of Medicine, Macau University of Science and Technology, Macau 999078, China; State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai 201203, China; Department of Pharmacology, Guilin Medical University, Guilin 541004, China.
Organizational Affiliation: