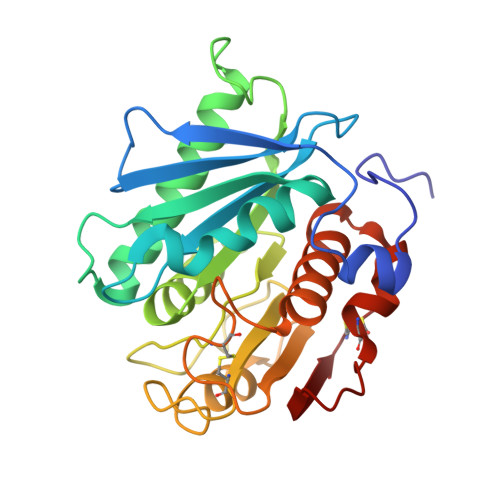
Complete decomposition of poly(ethylene terephthalate) by crude PET hydrolytic enzyme produced in Pichia pastoris
Chen, C.C., Li, X., Min, J., Zeng, Z., Ning, Z., He, H., Long, X., Niu, D., Peng, R., Liu, X., Yang, Y., Huang, J.W., Guo, R.T.(2023) Chem Eng J : 148418