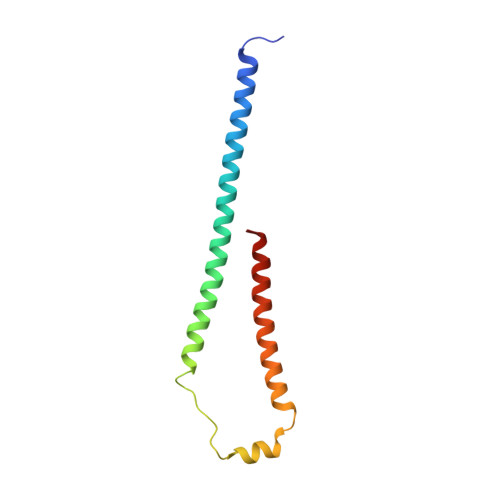
Cryo-EM structure of a bacteriophage M13 mini variant.
Jia, Q., Xiang, Y.(2023) Nat Commun 14: 5421-5421
- PubMed: 37669979
- DOI: https://doi.org/10.1038/s41467-023-41151-7
- Primary Citation of Related Structures:
8IXJ, 8IXK, 8IXL, 8JWT - PubMed Abstract:
Filamentous bacteriophages package their circular, single stranded DNA genome with the major coat protein pVIII and the minor coat proteins pIII, pVII, pVI, and pIX. Here, we report the cryo-EM structure of a ~500 Å long bacteriophage M13 mini variant. The distal ends of the mini phage are sealed by two cap-like complexes composed of the minor coat proteins. The top cap complex consists of pVII and pIX, both exhibiting a single helix structure. Arg33 of pVII and Glu29 of pIX, located on the inner surface of the cap, play a key role in recognizing the genome packaging signal. The bottom cap complex is formed by the hook-like structures of pIII and pVI, arranged in helix barrels. Most of the inner ssDNA genome adopts a double helix structure with a similar pitch to that of the A-form double-stranded DNA. These findings provide insights into the assembly of filamentous bacteriophages.
- Beijing Frontier Research Center for Biological Structure, Center for Infectious Disease Research, SXMU-Tsinghua Collaborative Innovation Center for Frontier Medicine, Department of Basic Medical Sciences, School of Medicine, Tsinghua University, Beijing, 100084, P.R. China.
Organizational Affiliation: