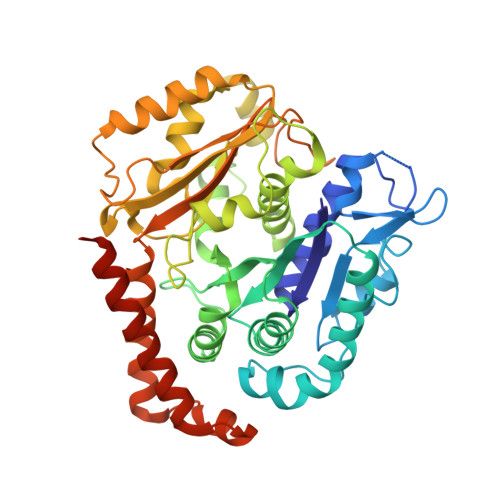
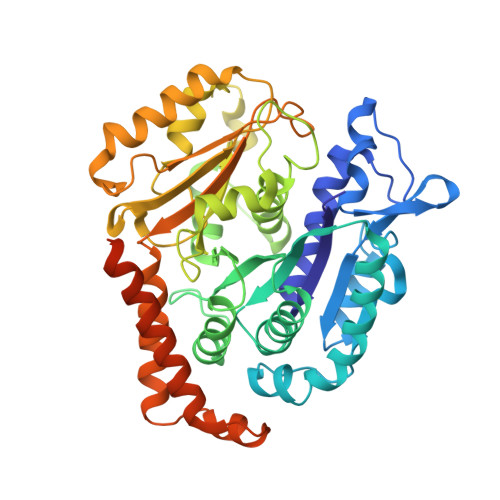
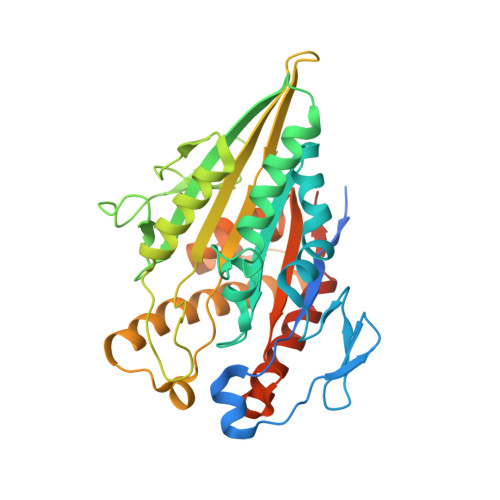
Cryo-EM of alpha-tubulin isotype-containing microtubules revealed a contracted structure of alpha 4A/ beta 2A microtubules.
Diao, L., Zheng, W., Zhao, Q., Liu, M., Fu, Z., Zhang, X., Bao, L., Cong, Y.(2023) Acta Biochim Biophys Sin (Shanghai) 55: 1551-1560
- PubMed: 37439022
- DOI: https://doi.org/10.3724/abbs.2023130
- Primary Citation of Related Structures:
8IXA, 8IXB, 8IXD, 8IXE, 8IXF, 8IXG - PubMed Abstract:
Microtubules are hollow α/β-tubulin heterodimeric polymers that play critical roles in cells. In vertebrates, both α- and β-tubulins have multiple isotypes encoded by different genes, which are intrinsic factors in regulating microtubule functions. However, the structures of microtubules composed of different tubulin isotypes, especially α-tubulin isotypes, remain largely unknown. Here, we purify recombinant tubulin heterodimers composed of different mouse α-tubulin isotypes, including α1A, α1C and α4A, with the β-tubulin isotype β2A. We further assemble and determine the cryo-electron microscopy (cryo-EM) structures of α1A/β2A, α1C/β2A, and α4A/β2A microtubules. Our structural analysis demonstrates that α4A/β2A microtubules exhibit longitudinal contraction between tubulin interdimers compared with α1A/β2A and α1C/β2A microtubules. Collectively, our findings reveal that α-tubulin isotype composition can tune microtubule structures, and also provide evidence for the "tubulin code" hypothesis.
- State Key Laboratory of Cell Biology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, University of Chinese Academy of Sciences, Chinese Academy of Sciences, Shanghai 200031, China.
Organizational Affiliation: