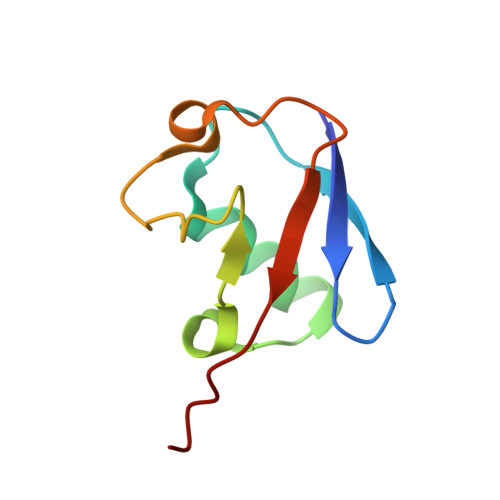
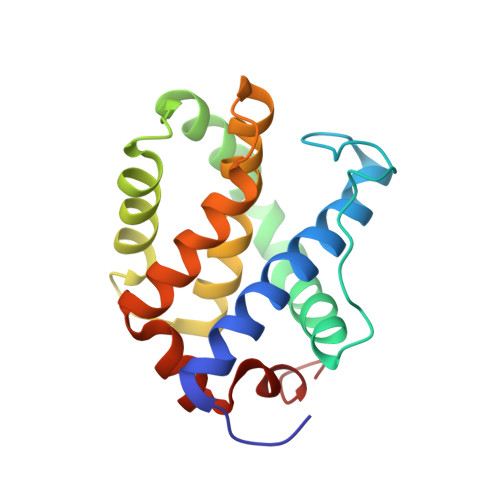
Parkin-mediated ubiquitination inhibits BAK apoptotic activity by blocking its canonical hydrophobic groove.
Cheng, P., Hou, Y., Bian, M., Fang, X., Liu, Y., Rao, Y., Cao, S., Liu, Y., Zhang, S., Chen, Y., Dong, X., Liu, Z.(2023) Commun Biol 6: 1260-1260
- PubMed: 38087033
- DOI: https://doi.org/10.1038/s42003-023-05650-z
- Primary Citation of Related Structures:
8IVB - PubMed Abstract:
BAK permeabilizes the mitochondrial outer membrane, causing apoptosis. This apoptotic activity of BAK is stimulated by binding prodeath activators within its canonical hydrophobic groove. Parkin, an E3 ubiquitin (Ub) ligase, can ubiquitinate BAK, which inhibits BAK apoptotic activity. However, the molecular mechanism underlying the inhibition of ubiquitination remains structurally uncharacterized. Here, we utilize truncated and soluble BAK to construct a mimetic of K113-ubiquitinated BAK (disulfide-linked Ub G76C ~ BAK K113C ) and further present its NMR-derived structure model. The classical L8-I44-H68-V70 hydrophobic patch of the conjugated Ub subunit binds within the canonical hydrophobic groove of BAK. This Ub occludes the binding of prodeath BID activators in the groove and impairs BID-triggered BAK activation and membrane permeabilization. Reduced interaction between Ub and BAK subunits allows BID to activate K113-ubiquitinated BAK. These mechanistic insights suggest a nonsignaling function of Ub in that it directly antagonizes stimuli targeting Ub-modified proteins rather than by recruiting downstream partners for cellular messaging.
- National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, 430070, China.
Organizational Affiliation: